7XD2
 
 | |
8JZN
 
 | | Structure of a fungal 1,3-beta-glucan synthase | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, C, You, Z, Chen, D, Hang, J, Wang, Z, Meng, J, Wang, L, Zhao, P, Qiao, J, Yun, C, Bai, L. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure of a fungal 1,3-beta-glucan synthase.
Sci Adv, 9, 2023
|
|
6U6Y
 
 | | Human SAMHD1 bound to ribo(CGCCU)-oligonucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, RNA CGCCU, ... | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
1VMP
 
 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
8JQ9
 
 | | Structure of Gabija GajA | | Descriptor: | Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8JQC
 
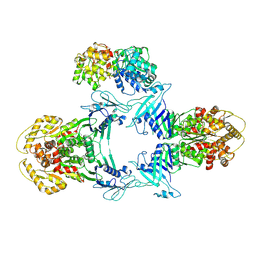 | | Structure of Gabija GajA-GajB 4:1 complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8JQB
 
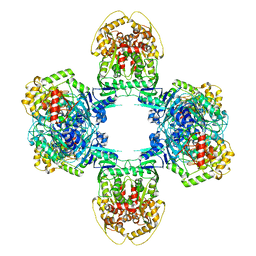 | | Structure of Gabija GajA-GajB 4:4 Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
1SQW
 
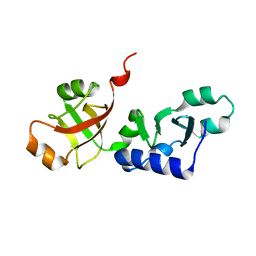 | | Crystal structure of KD93, a novel protein expressed in the human pro | | Descriptor: | Saccharomyces cerevisiae Nip7p homolog | | Authors: | Liu, J.F, Wang, X.Q, Wang, Z.X, Chen, J.R, Jiang, T, An, X.M, Chan, W.R, Liang, D.C. | | Deposit date: | 2004-03-19 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of KD93, a novel protein expressed in human hematopoietic stem/progenitor cells.
J.Struct.Biol., 148, 2004
|
|
6UK5
 
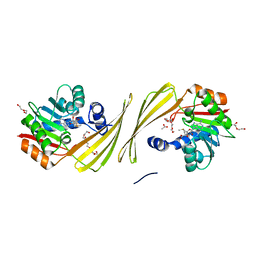 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
6O9H
 
 | | Mouse ECD with Fab1 | | Descriptor: | Gastric inhibitory polypeptide receptor, Heavy chain, Light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
8GU5
 
 | | Wild type poly(ethylene terephthalate) hydrolase | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
8GU4
 
 | | Poly(ethylene terephthalate) hydrolase (IsPETase)-linker | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
8K3S
 
 | | Structure of PKD2-F604P complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4TTH
 
 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
4CSU
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
4YNZ
 
 | | Structure of the N-terminal domain of SAD | | Descriptor: | Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4YOM
 
 | | Structure of SAD kinase | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
1LQX
 
 | | Crystal structure of V45E mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-14 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LR6
 
 | | Crystal structure of V45Y mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-15 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8IUI
 
 | |
1J55
 
 | | The Crystal Structure of Ca+-bound Human S100P Determined at 2.0A Resolution by X-ray | | Descriptor: | CALCIUM ION, S-100P PROTEIN | | Authors: | Zhang, H, Wang, G, Ding, Y, Wang, Z, Barraclough, R, Rudland, P.S, Fernig, D.G, Rao, Z. | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure at 2A Resolution of the Ca2+-binding Protein S100P
J.Mol.Biol., 325, 2003
|
|
6CXC
 
 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
