7WO1
 
 | | Discovery of SARS-CoV-2 3CLpro peptidomimetic inhibitors through H41-specific protein-ligand interactions | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
4PF7
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
8XAU
 
 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAV
 
 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8HY7
 
 | | EGFR kinase domain mutant "TMLR" with compound 28f | | Descriptor: | Epidermal growth factor receptor, ~{N}-[3-[[6-fluoranyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]quinazolin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The crystal structure of EGFR kinase domain(T790M/L858R) in complex with compound 28f
To Be Published
|
|
8U2B
 
 | |
6AE8
 
 | | Structure insight into histone chaperone Chz1-mediated H2A.Z recognition and replacement | | Descriptor: | BICINE, Histone H2A.Z-specific chaperone CHZ1, Histone H2B.1,Histone H2A.Z | | Authors: | Wang, Y.Y, Shan, S, Zhou, Z. | | Deposit date: | 2018-08-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement.
Plos Biol., 17, 2019
|
|
8IOG
 
 | |
2KKR
 
 | | Solution structure of SCA7 zinc finger domain from human ataxin-7 protein | | Descriptor: | Ataxin-7, ZINC ION | | Authors: | Wang, Y, Atkinson, A.R, Bonnet, J, Romier, C, Kieffer, B, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Histone deubiquitination by SAGA is modulated by an atypical zinc finger domain of Ataxin-7
To be Published
|
|
5XYX
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5XYY
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5ZQ3
 
 | | PDE-Ubiquitin | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ2
 
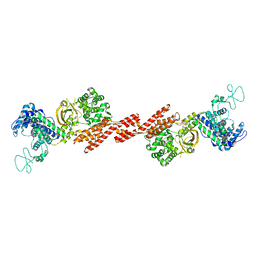 | | SidE apo form | | Descriptor: | SidE | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ6
 
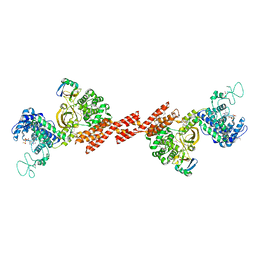 | | SidE-Ubi-ADPr | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
8HXB
 
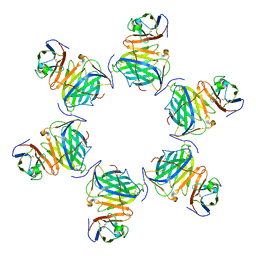 | | Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXC
 
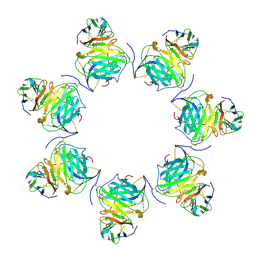 | | Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXA
 
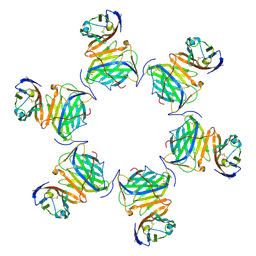 | | Cryo-EM structure of MPXV M2 in complex with human B7.1 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD80 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HYN
 
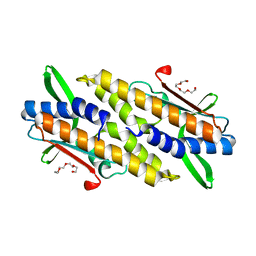 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWJ
 
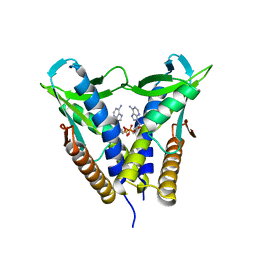 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
7DMZ
 
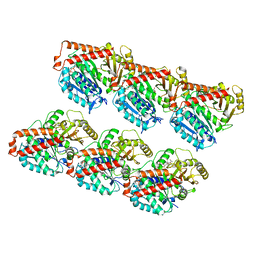 | | GMPCPP microtubule complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Wang, Y.X. | | Deposit date: | 2020-12-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | GMPCPP microtubule complex
To Be Published
|
|
7DN0
 
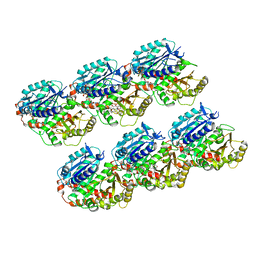 | | AJ-GMPCPP-MT-non-seam | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Wang, Y.X. | | Deposit date: | 2020-12-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | GMPCPP microtubule
To Be Published
|
|
5ZQ5
 
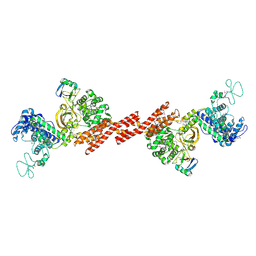 | | SidE-Ubi | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ4
 
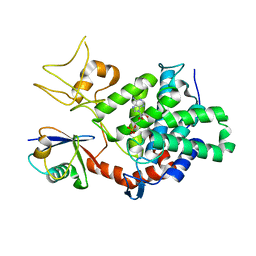 | | PDE-Ubi-ADPr | | Descriptor: | ADENOSINE MONOPHOSPHATE, SidE, ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5XYZ
 
 | | The structure of human BTK kinase domain in complex with a covalent inhibitor | | Descriptor: | N-[3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-1H-1,2,4-triazol-3-yl)phenyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Xie, Y.T, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
