1ZMH
 
 | | Crystal structure of human neutrophil peptide 2, HNP-2 (variant Gly16-> D-Ala) | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, Neutrophil defensin 2, ... | | Authors: | Lubkowski, J, Prahl, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reconstruction of the conserved beta-bulge in mammalian defensins using D-amino acids.
J.Biol.Chem., 280, 2005
|
|
1UDE
 
 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
3M4V
 
 | | Crystal structure of the A330P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-03-12 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3)
Chembiochem, 11, 2010
|
|
6MKS
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | Chimera protein of NLR family CARD domain-containing protein 4 and EGFP | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NLRC4CARDfilament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly.
J. Biol. Chem., 293, 2018
|
|
4R3Q
 
 | | Crystal structure of SYCE3 | | Descriptor: | Synaptonemal complex central element protein 3 | | Authors: | Lu, J, Feng, J, Zhou, W, Yang, X, Shen, Y. | | Deposit date: | 2014-08-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural insight into the central element assembly of the synaptonemal complex
Sci Rep, 4, 2014
|
|
5SWF
 
 | | The structure of the PP2A B56 subunit double phosphorylated BubR1 complex | | Descriptor: | Double phosphorylated BubR1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Page, R, Wang, X, Bajaj, R, Peti, W. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Expanding the PP2A Interactome by Defining a B56-Specific SLiM.
Structure, 24, 2016
|
|
5TUC
 
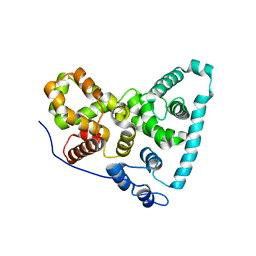 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | Descriptor: | Sus TBC1D15 GAP Domain | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
2HXU
 
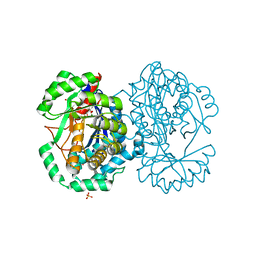 | | Crystal structure of K220A mutant of L-Fuconate Dehydratase from Xanthomonas campestris liganded with Mg++ and L-fuconate | | Descriptor: | 6-deoxy-L-galactonic acid, L-fuconate dehydratase, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: l-Fuconate Dehydratase from Xanthomonas campestris.
Biochemistry, 45, 2006
|
|
4QV4
 
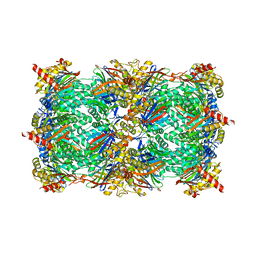 | | yCP beta5-M45T mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
1YJX
 
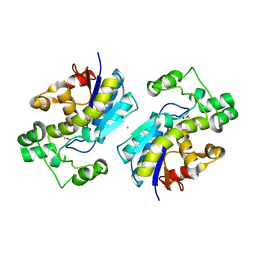 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, CITRIC ACID, Phosphoglycerate mutase 1 | | Authors: | Wang, Y, Wei, Z, Liu, L, Gong, W. | | Deposit date: | 2005-01-16 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human B-type phosphoglycerate mutase bound with citrate.
Biochem.Biophys.Res.Commun., 331, 2005
|
|
1YAX
 
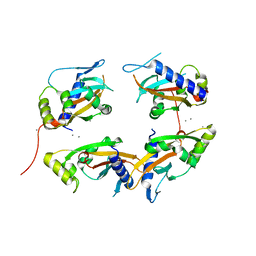 | |
4QW6
 
 | | yCP beta5-M45V mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
3M9W
 
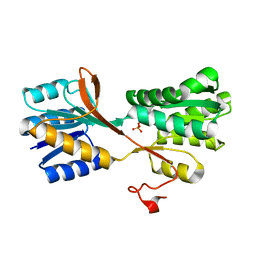 | |
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4RC2
 
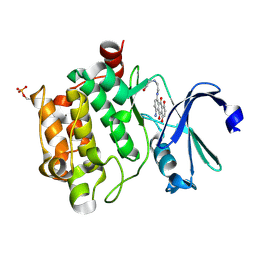 | |
1U1D
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(phenylthio)acyclouridine (ptau) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLTHIO)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U39
 
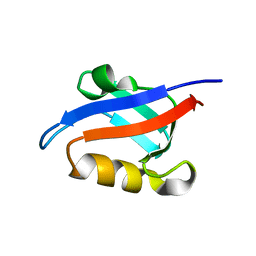 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1BSV
 
 | | GDP-FUCOSE SYNTHETASE FROM ESCHERICHIA COLI COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-FUCOSE SYNTHETASE) | | Authors: | Somers, W.S, Stahl, M.L, Sullivan, F.X. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site.
Structure, 6, 1998
|
|
4QQ4
 
 | | CW-type zinc finger of MORC3 in complex with the amino terminus of histone H3 | | Descriptor: | CHLORIDE ION, Histone H3.3, MORC family CW-type zinc finger protein 3, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
1YHN
 
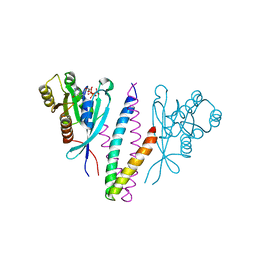 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
3MOK
 
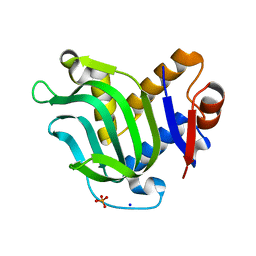 | | Structure of Apo HasAp from Pseudomonas aeruginosa to 1.55A Resolution | | Descriptor: | Heme acquisition protein HasAp, PHOSPHATE ION, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
4P5A
 
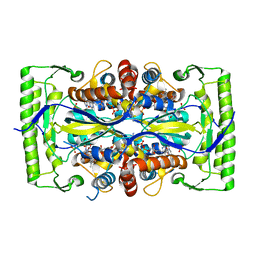 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
5UB9
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni | | Descriptor: | ACETATE ION, ATP phosphoribosyltransferase, CHLORIDE ION, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
