2CIY
 
 | | Chloroperoxidase complexed with cyanide and DMSO | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
2X3W
 
 | | structure of mouse syndapin I (crystal form 2) | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 1 | | Authors: | Ma, Q, Rao, Y, Saenger, W, Haucke, V. | | Deposit date: | 2010-01-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular Basis for SH3 Domain Regulation of F-Bar-Mediated Membrane Deformation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
2CJ0
 
 | | chloroperoxidase complexed with nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
5HTP
 
 | |
2C9Z
 
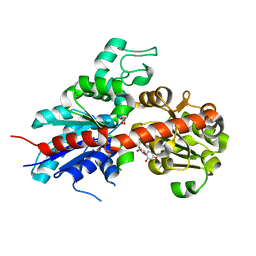 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2CEH
 
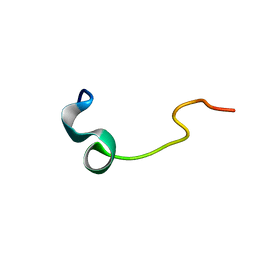 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
5HBP
 
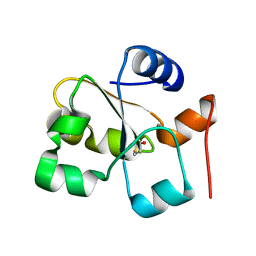 | | The crystal of rhodanese domain of YgaP treated with SNOC | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBU
 
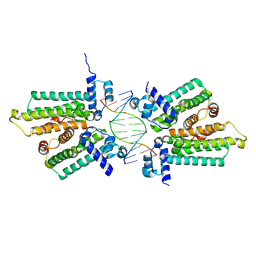 | |
2CAX
 
 | | STRUCTURAL BASIS FOR COOPERATIVE BINDING OF RIBBON-HELIX-HELIX REPRESSOR OMEGA TO MUTATED DIRECT DNA HEPTAD REPEATS | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*CP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*GP *TP*CP*AP*CP*AP*AP*GP*C)-3', ... | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-12-23 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
2CJ1
 
 | | chloroperoxidase complexed with formate (ethylene glycol cryoprotectant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
2CB5
 
 | | HUMAN BLEOMYCIN HYDROLASE, C73S/DELE455 MUTANT | | Descriptor: | PROTEIN (BLEOMYCIN HYDROLASE) | | Authors: | O'Farrell, P.A, Gonzalez, F, Zheng, W, Johnston, S.A, Joshua-Tor, L. | | Deposit date: | 1999-03-02 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human bleomycin hydrolase, a self-compartmentalizing cysteine protease.
Structure Fold.Des., 7, 1999
|
|
2CP9
 
 | | Solution structure of RSGI RUH-042, a UBA domain from human mitochondrial elongation factor Ts | | Descriptor: | Elongation factor Ts, mitochondrial | | Authors: | Ohashi, W, Hirota, H, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-042, a UBA domain from human mitochondrial elongation factor Ts
To be Published
|
|
7RRL
 
 | |
1FXU
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CALF SPLEEN IN COMPLEX WITH N(7)-ACYCLOGUANOSINE INHIBITOR AND A PHOSPHATE ION | | Descriptor: | 2-AMINO-7-[2-(2-HYDROXY-1-HYDROXYMETHYL-ETHYLAMINO)-ETHYL]-1,7-DIHYDRO-PURIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Luic, M, Bzowska, A, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase: structure of its ternary complex with an N(7)-acycloguanosine inhibitor and a phosphate anion.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1A9E
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-05 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
5JBW
 
 | | Crystal structure of LiuC | | Descriptor: | 3-hydroxybutyryl-CoA dehydratase | | Authors: | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-04-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
4YJY
 
 | |
2CBN
 
 | |
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
2CEF
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity. | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
2CH6
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with ADP and glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-ACETYL-D-GLUCOSAMINE KINASE, alpha-D-glucopyranose | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
5J19
 
 | | phospho-Pon binding-induced Plk1 dimerization | | Descriptor: | GLYCEROL, Phosphorylated peptide from Partner of Numb, Serine/threonine-protein kinase PLK1 | | Authors: | Zhu, K, Shan, Z, Zhang, L, Wen, W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phospho-Pon Binding-Mediated Fine-Tuning of Plk1 Activity
Structure, 24, 2016
|
|
2B5D
 
 | | Crystal structure of the novel alpha-amylase AmyC from Thermotoga maritima | | Descriptor: | alpha-Amylase | | Authors: | Dickmanns, A, Ballschmiter, M, Liebl, W, Ficner, R. | | Deposit date: | 2005-09-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the novel alpha-amylase AmyC from Thermotoga maritima.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2UWD
 
 | | Inhibition of the HSP90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole, isoxazole amide analogs | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION, ... | | Authors: | Sharp, S.Y, Prodromou, C, Boxall, K, Powers, M.V, Holmes, J.L, Box, G, Matthews, T.P, Cheung, K.M, Kalusa, A, James, K, Hayes, A, Hardcastle, A, Dymock, B, Brough, P.A, Barril, X, Cansfield, J.E, Wright, L.M, Surgenor, A, Foloppe, N, Aherne, W, Pearl, L, Jones, K, McDonald, E, Raynaud, F, Eccles, S, Drysdale, M, Workman, P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of the heat shock protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues.
Mol. Cancer Ther., 6, 2007
|
|
