7CLU
 
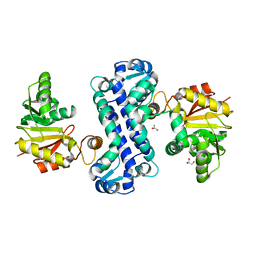 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
3O40
 
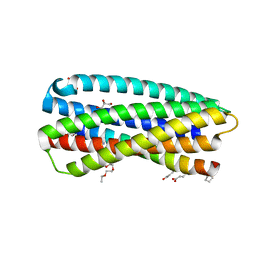 | | Complex of a chimeric alpha/beta-peptide based on the gp41 CHR domain bound to gp41-5 | | Descriptor: | CHLORIDE ION, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2010-07-26 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
3O42
 
 | |
2MHU
 
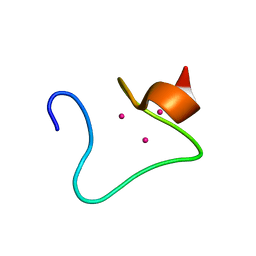 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
3O3Z
 
 | |
4LFM
 
 | |
1RJ5
 
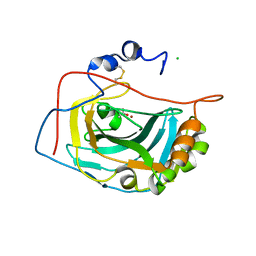 | | Crystal Structure of the Extracellular Domain of Murine Carbonic Anhydrase XIV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Whittington, D.A, Grubb, J.H, Waheed, A, Shah, G.N, Sly, W.S, Christianson, D.W. | | Deposit date: | 2003-11-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
J.Biol.Chem., 279, 2004
|
|
3O3X
 
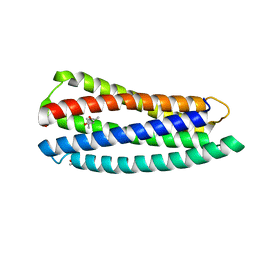 | | Crystal structure of gp41-5, a single-chain 5-helix-bundle based on HIV gp41 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, gp41-5 | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2010-07-26 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
1U9G
 
 | | Heterocyclic Peptide Backbone Modification in GCN4-pLI Based Coiled Coils: Replacement of K(8)L(9) | | Descriptor: | General control protein GCN4, SULFATE ION | | Authors: | Horne, W.S, Yadav, M.K, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-09 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterocyclic peptide backbone modifications in an alpha-helical coiled coil.
J.Am.Chem.Soc., 126, 2004
|
|
1U9H
 
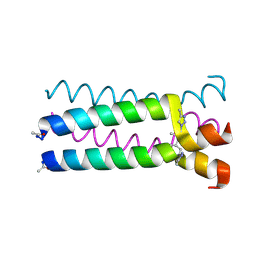 | |
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4LFN
 
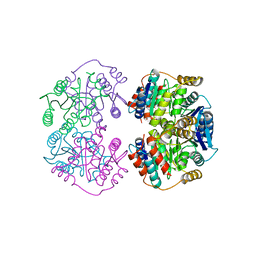 | |
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | Descriptor: | RNA-binding protein 3 | | Authors: | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
3NIL
 
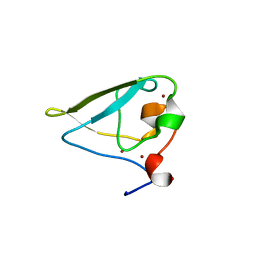 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIJ
 
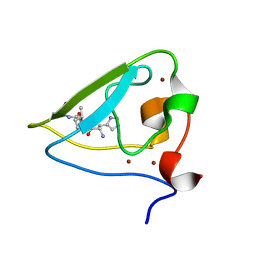 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
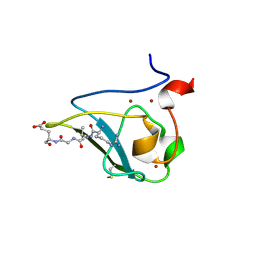 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
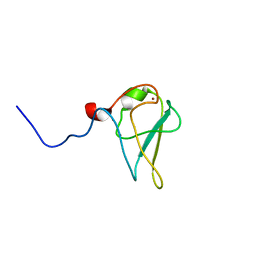 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
4LFK
 
 | |
1SO6
 
 | | Crystal structure of E112Q/H136A double mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
4LFL
 
 | |
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
6BII
 
 | | Crystal Structure of Pyrococcus yayanosii Glyoxylate Hydroxypyruvate Reductase in complex with NADP and malonate (re-refinement of 5AOW) | | Descriptor: | GLYCEROL, Glyoxylate reductase, MALONATE ION, ... | | Authors: | Lassalle, L, Shabalin, I.G, Girard, E, Minor, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.
Sci Rep, 6, 2016
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
