3ZW9
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA | | Descriptor: | (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
1OJG
 
 | | Sensory domain of the membraneous two-component fumarate sensor DcuS of E. coli | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Pappalardo, L, Janausch, I.G, Vijayan, V, Zientz, E, Junker, J, Peti, W, Zweckstetter, M, Unden, G, Griesinger, C. | | Deposit date: | 2003-07-10 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli.
J. Biol. Chem., 278, 2003
|
|
8AB9
 
 | | Complex III2 from Yarrowia lipolytica, ascorbate-reduced, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AB7
 
 | | Complex III2 from Yarrowia lipolytica, atovaquone and antimycin A bound | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AB6
 
 | | Complex III2 from Yarrowia lipolytica, combined datasets, consensus refinement | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABB
 
 | | Complex III2 from Yarrowia lipolytica, ascorbate-reduced, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABJ
 
 | | Complex III2 from Yarrowia lipolytica, antimycin A bound, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABM
 
 | | Complex III2 from Yarrowia lipolytica, apo, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC4
 
 | | Complex III2 from Yarrowia lipolytica, apo, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
8ABA
 
 | | Complex III2 from Yarrowia lipolytica, ascorbate-reduced, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABE
 
 | | Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
1OR4
 
 | |
8ABH
 
 | | Complex III2 from Yarrowia lipolytica, antimycin A bound, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC3
 
 | | Complex III2 from Yarrowia lipolytica, apo, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
3EYV
 
 | | Anti-Lewis Y Fab fragment with Lewis Y antigen in the presence of zinc ions | | Descriptor: | GLYCEROL, ZINC ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Farrugia, W, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A possible role for metallic ions in the carbohydrate cluster recognition displayed by a lewis y specific antibody.
Plos One, 4, 2009
|
|
8ABF
 
 | | Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
3ZLA
 
 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
8ABK
 
 | | Complex III2 from Yarrowia lipolytica, decylubiquinol bound, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AB8
 
 | | Complex III2, b-position, with decylubiquinone and ascorbate-reduced | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
3ZIM
 
 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | Descriptor: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
3KEM
 
 | | Crystal structure of IspH:IPP complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5BWL
 
 | |
2IS6
 
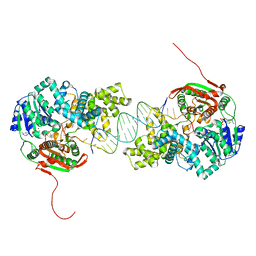 | | Crystal structure of UvrD-DNA-ADPMgF3 ternary complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*GP*TP*TP*AP*T)-3', ADENOSINE-5'-DIPHOSPHATE, DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
3KE9
 
 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
