3RTE
 
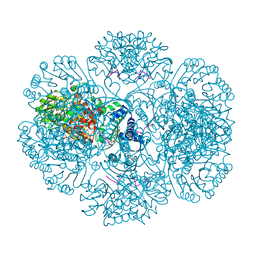 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP and ATP. | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-03 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4BSU
 
 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in C2 crystal form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | 著者 | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | 登録日 | 2013-06-11 | | 公開日 | 2013-06-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4CD2
 
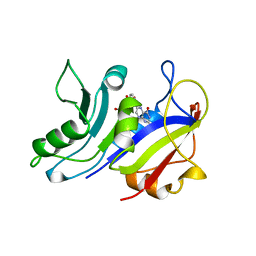 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | 分子名称: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | 著者 | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Pangborn, W, Queener, S.F. | | 登録日 | 1999-03-18 | | 公開日 | 2000-03-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
3RNO
 
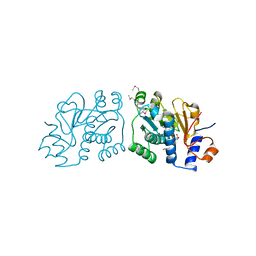 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with NADP. | | 分子名称: | Apolipoprotein A-I-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-22 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROZ
 
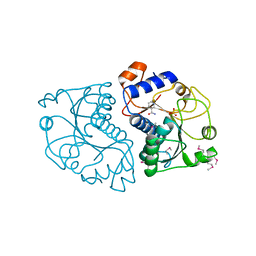 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | 分子名称: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-26 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | 分子名称: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | 著者 | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-04-27 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P3-Di(adenosine-5') triphosphate | | 分子名称: | BIS(ADENOSINE)-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-02 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSQ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-02 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.054 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-03 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RU9
 
 | |
3RUH
 
 | |
3RU7
 
 | |
8VGR
 
 | |
8VJS
 
 | |
4MOV
 
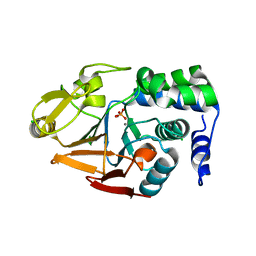 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2013-09-12 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4503 Å) | | 主引用文献 | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4E3E
 
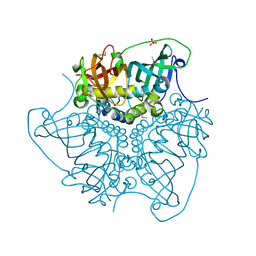 | | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl | | 分子名称: | MaoC domain protein dehydratase, SULFATE ION | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-09 | | 公開日 | 2012-03-21 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
8VJR
 
 | |
4DE9
 
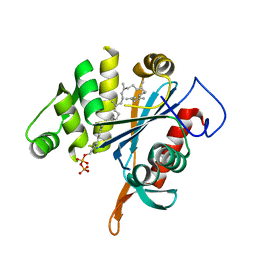 | | LytR-CPS2A-psr family protein YwtF (TagT) with bound octaprenyl pyrophosphate lipid | | 分子名称: | (2Z,6Z,10Z,14Z,18Z,22E,26E)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, Putative transcriptional regulator ywtF | | 著者 | Eberhardt, A, Hoyland, C.N, Vollmer, D, Bisle, S, Cleverley, R.M, Johnsborg, O, Havarstein, S, Lewis, R.J, Vollmer, W. | | 登録日 | 2012-01-20 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.787 Å) | | 主引用文献 | Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae.
Microb Drug Resist, 18, 2012
|
|
3RUD
 
 | |
8X1C
 
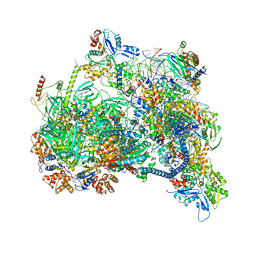 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
4MUY
 
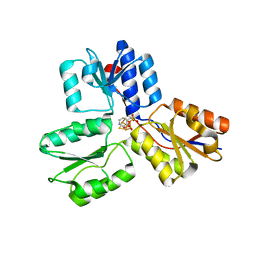 | | IspH in complex with pyridin-4-ylmethyl diphosphate | | 分子名称: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-4-ylmethyl trihydrogen diphosphate | | 著者 | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | 登録日 | 2013-09-23 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
3RXW
 
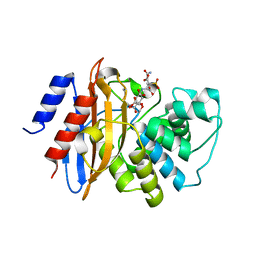 | | KPC-2 carbapenemase in complex with PSR3-226 | | 分子名称: | (2S,3R)-4-(2-amino-2-oxoethoxy)-3-(dihydroxy-lambda~4~-sulfanyl)-3-methyl-4-oxo-2-{[(1E)-3-oxoprop-1-en-1-yl]amino}butanoic acid, CITRIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | 著者 | Ke, W, van den Akker, F. | | 登録日 | 2011-05-10 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
3RU3
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH and ATP. | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, MAGNESIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-04 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.605 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4MMH
 
 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
8VZ8
 
 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | 分子名称: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Ciacchi, L, Rossjohn, J, Awad, W. | | 登録日 | 2024-02-11 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
