5HNF
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*(YPE)P*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*(YPF)P*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
5HTN
 
 | |
3TFZ
 
 | | Crystal structure of Zhui aromatase/cyclase from Streptomcyes sp. R1128 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclase, POTASSIUM ION | | Authors: | Ames, B.D, Lee, M.Y, Moody, C, Zhang, W, Tang, Y, Wong, S.K, Tsai, S.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Characterization of ZhuI Aromatase/Cyclase from the R1128 Polyketide Pathway.
Biochemistry, 50, 2011
|
|
3SPW
 
 | | Structure of Osh4p/Kes1p in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Protein KES1 | | Authors: | Delfosse, V, de Saint-Jean, M, Douguet, D, Antonny, B, Drin, G, Bourguet, W. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers.
J.Cell Biol., 195, 2011
|
|
4NPM
 
 | |
3SOV
 
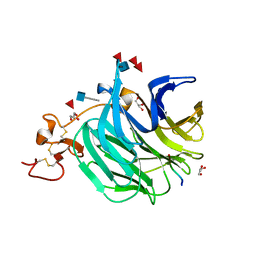 | | The structure of a beta propeller domain in complex with peptide S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
5HOZ
 
 | |
4NS5
 
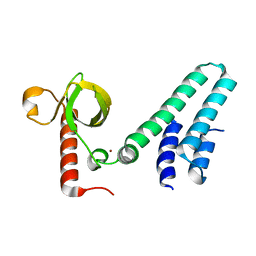 | | Crystal structure of human BS69 Bromo-Zinc finger-PWWP | | Descriptor: | ZINC ION, Zinc finger MYND domain-containing protein 11 | | Authors: | Wang, J.C, Qin, S, Li, F.D, Li, S, Zhang, W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36me3 nucleosome binding.
Cell Res., 24, 2014
|
|
3SSP
 
 | |
4ND9
 
 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
3SSV
 
 | |
5HWP
 
 | | MvaS with acetylated Cys115 in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWZ
 
 | | Crystal structure of nitrophorin 4 D30N mutant with nitrite | | Descriptor: | NITRITE ION, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ogata, H, He, C, Lubitz, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the heme active site electronic structure affecting the unprecedented nitrite dismutase activity of the ferrihemebproteins, the nitrophorins.
Chem Sci, 7, 2016
|
|
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
4NWQ
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group F4132 | | Descriptor: | Putative uncharacterized protein PH0671, SULFATE ION, Uncharacterized protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
5HP7
 
 | | Crystal structures of RidA in the apo form | | Descriptor: | Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
5HPA
 
 | |
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
5HWO
 
 | | MvaS in complex with 3-hydroxy-3-methylglutaryl coenzyme A | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
4NHD
 
 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
3T7B
 
 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
4NPA
 
 | | Scrystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Dorovatovsky, P.V, Lipkin, A.V, Minor, W, Shumilin, I.A, Popov, V.O. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup
To be Published
|
|
3TAP
 
 | |
4NW2
 
 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
