3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2PRJ
 
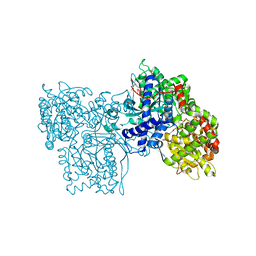 | | Binding of N-acetyl-beta-D-glucopyranosylamine to Glycogen Phosphorylase B | | Descriptor: | GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kontou, M, Zographos, S.E, Watson, K.A, Johnson, L.N, Bichard, C.J.F, Fleet, G.W.J, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-acetyl-beta-D-glucopyranosylamine: a potent T-state inhibitor of glycogen phosphorylase. A comparison with alpha-D-glucose.
Protein Sci., 4, 1995
|
|
1PBF
 
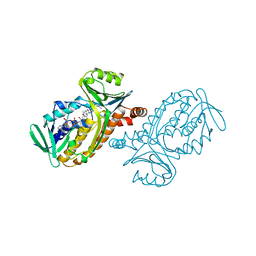 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PDH
 
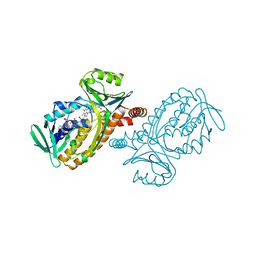 | |
1PBC
 
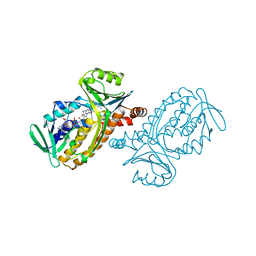 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
2UU8
 
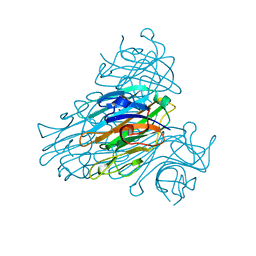 | | X-ray structure of Ni, Ca concanavalin A at Ultra-high resolution (0. 94A) | | Descriptor: | CALCIUM ION, CONCANAVALIN, NICKEL (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-01 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UUJ
 
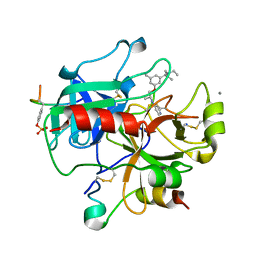 | | Thrombin-hirugen-gw473178 ternary complex at 1.32A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UUK
 
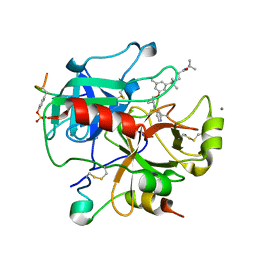 | | Thrombin-hirugen-gw420128 ternary complex at 1.39A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UUF
 
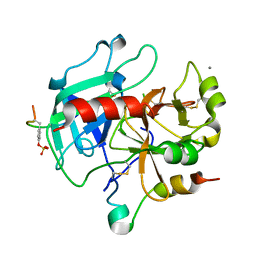 | | Thrombin-hirugen binary complex at 1.26A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3TOR
 
 | | Crystal structure of Escherichia coli NrfA with Europium bound | | Descriptor: | CALCIUM ION, Cytochrome c nitrite reductase, EUROPIUM ION, ... | | Authors: | Lockwood, C.W.J, Clarke, T.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Biochem.Soc.Trans., 39, 2011
|
|
1FX7
 
 | | CRYSTAL STRUCTURE OF THE IRON-DEPENDENT REGULATOR (IDER) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | COBALT (II) ION, IRON-DEPENDENT REPRESSOR IDER, SULFATE ION | | Authors: | Feese, M.D, Ingason, B.P, Goranson-Siekierke, J, Holmes, R.K, Hol, W.J.G. | | Deposit date: | 2000-09-25 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the iron-dependent regulator from Mycobacterium tuberculosis at 2.0-A resolution reveals the Src homology domain 3-like fold and metal binding function of the third domain.
J.Biol.Chem., 276, 2001
|
|
1XD3
 
 | | Crystal structure of UCHL3-UbVME complex | | Descriptor: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | Authors: | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
1DZN
 
 | | Asp170Ser mutant of vanillyl-alcohol oxidase | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, FLAVIN-ADENINE DINUCLEOTIDE, VANILLYL-ALCOHOL OXIDASE | | Authors: | Van Den heuvel, R.H.H, Fraaije, M.W, Van Berkel, W.J.H, Mattevi, A. | | Deposit date: | 2000-03-05 | | Release date: | 2000-03-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asp170 is Crucial for the Redox Properties of Vanillyl-Alcohol Oxidase
J.Biol.Chem., 275, 2000
|
|
1E0Y
 
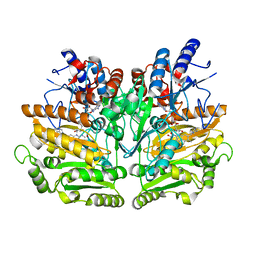 | | Structure of the D170S/T457E double mutant of vanillyl-alcohol oxidase | | Descriptor: | ALPHA,ALPHA,ALPHA-TRIFLUORO-P-CRESOL, FLAVIN-ADENINE DINUCLEOTIDE, VANILLYL-ALCOHOL OXIDASE | | Authors: | Van Der heuvel, R.H.H, Van Berkel, W.J.H, Mattevi, A. | | Deposit date: | 2000-04-11 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inversion of Stereospecificity in Vanillyl-Alcohol Oxidase
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ZAE
 
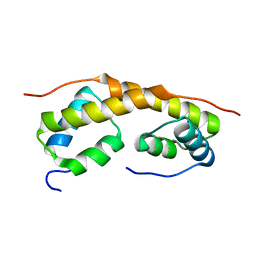 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
5TI3
 
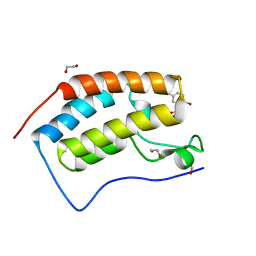 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 17503468 | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dibromo-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI2
 
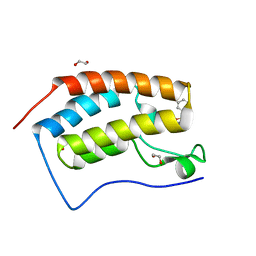 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 7635936 | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI7
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 17528462 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(3-(2-oxopyrrolidin-1-yl)phenyl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI6
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841881 | | Descriptor: | 1,2-ETHANEDIOL, 2,6-difluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
2GPN
 
 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
2GV4
 
 | | Solution structure of the poliovirus 3'-UTR Y-stem | | Descriptor: | Short RNA strand 5'-GGACCUCUCGAAAGAGUGGUCC-3' | | Authors: | Heus, H.A, Zoll, J, Tessari, M, van Kuppeveld, F.J.M, Melchers, W.J.G. | | Deposit date: | 2006-05-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Breaking pseudo-twofold symmetry in the poliovirus 3'-UTR Y-stem by restoring Watson-Crick base pairs.
Rna, 13, 2007
|
|
5TI5
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841880 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI4
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841871 | | Descriptor: | 1,2-ETHANEDIOL, 3-chloro-4-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
1SBS
 
 | | CRYSTAL STRUCTURE OF AN ANTI-HCG FAB | | Descriptor: | MONOCLONAL ANTIBODY 3A2, SULFATE ION | | Authors: | Fotinou, C, Beauchamp, J, Emsley, P, Dehaan, A, Schielen, W.J.G, Bos, E, Isaacs, N.W. | | Deposit date: | 1998-04-08 | | Release date: | 1999-04-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an Fab fragment against a C-terminal peptide of hCG at 2.0 A resolution.
J.Biol.Chem., 273, 1998
|
|
