3TI2
 
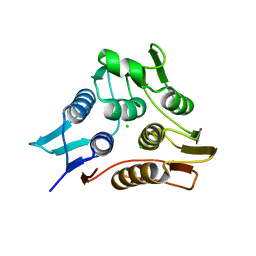 | | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-19 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae
TO BE PUBLISHED
|
|
3TNL
 
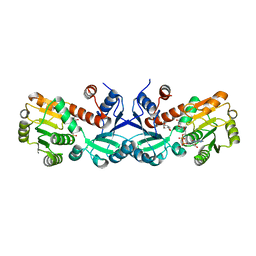 | | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD. | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.
TO BE PUBLISHED
|
|
3TOZ
 
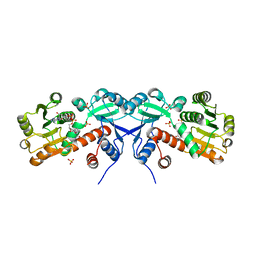 | | 2.2 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with NAD.
TO BE PUBLISHED
|
|
3TSB
 
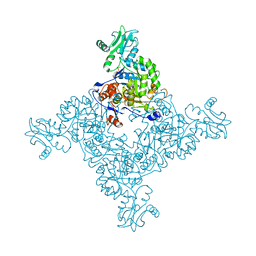 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
1ZTB
 
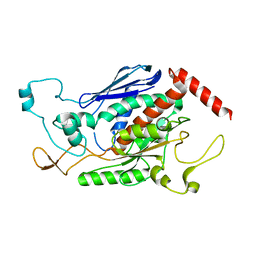 | | Crystal Structure of Chorismate Synthase from Mycobacterium tuberculosis | | Descriptor: | Chorismate synthase | | Authors: | Dias, M.V.B, Borges, J.C, Ely, F, Pereira, J.H, Canduri, F, Ramos, C.H.I, Frazzon, J, Palma, M.S, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of chorismate synthase from Mycobacterium tuberculosis
J.Struct.Biol., 154, 2006
|
|
3U9E
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. | | Descriptor: | ARGININE, CHLORIDE ION, COENZYME A, ... | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA.
To be Published
|
|
3TYS
 
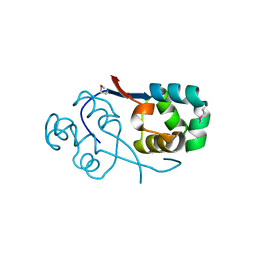 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3ZPD
 
 | |
1FLH
 
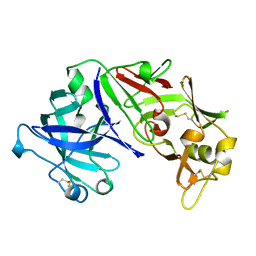 | | CRYSTAL STRUCTURE OF HUMAN UROPEPSIN AT 2.45 A RESOLUTION | | Descriptor: | UROPEPSIN | | Authors: | Canduri, F, Teodoro, L.G.V.L, Fadel, V, Lorenzi, C.C.B, Hial, V, Gomes, R.A.S, Neto, J.R, De Azevedo Jr, W.F. | | Deposit date: | 2000-08-14 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of human uropepsin at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FHJ
 
 | |
4IJC
 
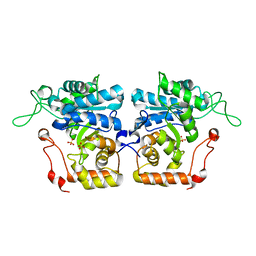 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IFA
 
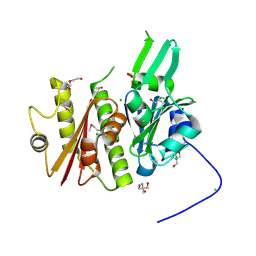 | | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Shatsman, S, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames
To be Published
|
|
4IJR
 
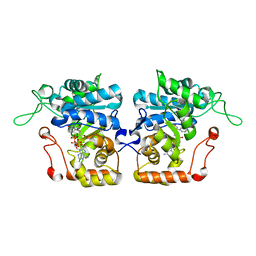 | | Crystal structure of Saccharomyces cerevisiae arabinose dehydrogenase Ara1 complexed with NADPH | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IIN
 
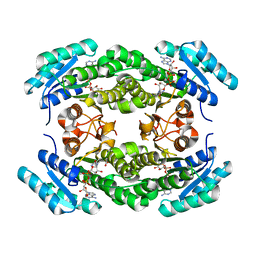 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+ | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
To be Published
|
|
4IJK
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), SODIUM ION | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I.G, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695
To be Published
|
|
4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4MPB
 
 | | 1.7 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus | | Descriptor: | Betaine aldehyde dehydrogenase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4IIU
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution
To be Published
|
|
4ICH
 
 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
4IIV
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.5 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.5 A resolution
To be Published
|
|
4IW7
 
 | | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis. | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Newcomb, W, Niedzialkowska, E, Porebski, P.J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis.
To be Published
|
|
4ISC
 
 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4ISX
 
 | | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, Maltose O-acetyltransferase | | Authors: | Tan, K, Gu, G, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa
To be Published
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
4IUO
 
 | | 1.8 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) K170M Mutant in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
