1GHX
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI7
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1G26
 
 | |
1G2A
 
 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | 分子名称: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | 著者 | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | 登録日 | 2000-10-18 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G3J
 
 | | CRYSTAL STRUCTURE OF THE XTCF3-CBD/BETA-CATENIN ARMADILLO REPEAT COMPLEX | | 分子名称: | BETA-CATENIN ARMADILLO REPEAT REGION, TCF3-CBD (CATENIN BINDING DOMAIN) | | 著者 | Graham, T.A, Weaver, C, Mao, F, Kimelman, D, Xu, W. | | 登録日 | 2000-10-24 | | 公開日 | 2000-12-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a beta-catenin/Tcf complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | 分子名称: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | 著者 | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | 登録日 | 2000-12-07 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
1FU1
 
 | | CRYSTAL STRUCTURE OF HUMAN XRCC4 | | 分子名称: | ACETIC ACID, DNA REPAIR PROTEIN XRCC4 | | 著者 | Junop, M, Modesti, M, Guarne, A, Gellert, M, Yang, W. | | 登録日 | 2000-09-13 | | 公開日 | 2000-12-11 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the Xrcc4 DNA repair protein and implications for end joining.
EMBO J., 19, 2000
|
|
1FUE
 
 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | 登録日 | 2000-09-15 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
1H3L
 
 | | N-terminal fragment of SigR from Streptomyces coelicolor | | 分子名称: | RNA POLYMERASE SIGMA FACTOR | | 著者 | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | 登録日 | 2002-09-10 | | 公開日 | 2002-10-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.375 Å) | | 主引用文献 | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
1H18
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | 分子名称: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | 著者 | Becker, A, Kabsch, W. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H16
 
 | |
1H17
 
 | |
1GPP
 
 | |
1GSW
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | 登録日 | 2002-01-09 | | 公開日 | 2002-02-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | 登録日 | 2002-01-09 | | 公開日 | 2002-02-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1EAW
 
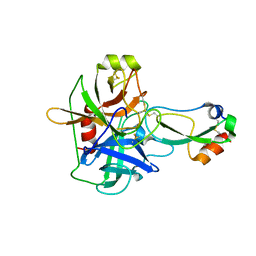 | |
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | 分子名称: | SYNDECAN-4 | | 著者 | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | 登録日 | 2000-03-04 | | 公開日 | 2001-03-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
1H8Z
 
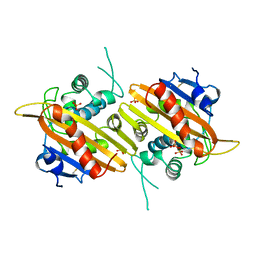 | | Crystal structure of the class D beta-lactamase OXA-13 | | 分子名称: | BETA-LACTAMASE, SULFATE ION | | 著者 | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | 登録日 | 2001-02-17 | | 公開日 | 2001-07-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
1HH5
 
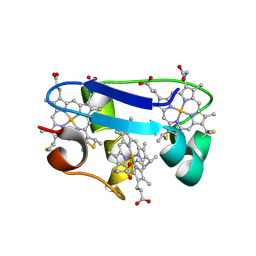 | | cytochrome c7 from Desulfuromonas acetoxidans | | 分子名称: | CYTOCHROME C7, HEME C | | 著者 | Czjzek, M, Haser, R, Arnoux, P, Shepard, W. | | 登録日 | 2000-12-21 | | 公開日 | 2001-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Cytochrome C7 from Desulfuromonas Acetoxidans at 1.9A Resolutio N
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1F2K
 
 | | CRYSTAL STRUCTURE OF ACANTHAMOEBA CASTELLANII PROFILIN II, CUBIC CRYSTAL FORM | | 分子名称: | PROFILIN II | | 著者 | Fedorov, A.A, Shi, W, Mahoney, N, Kaiser, D.A, Almo, S.C. | | 登録日 | 2000-05-26 | | 公開日 | 2000-06-08 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Comparative Structural Analysis of Profilins
To be Published
|
|
1F5S
 
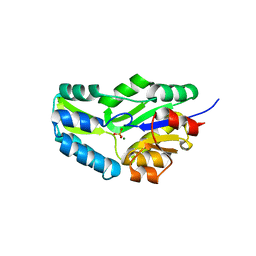 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | 著者 | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2000-06-15 | | 公開日 | 2001-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
1GSV
 
 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | 登録日 | 2002-01-08 | | 公開日 | 2002-02-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GYJ
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | 分子名称: | HYPOTHETICAL PROTEIN YDCE | | 著者 | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | 登録日 | 2002-04-23 | | 公開日 | 2002-10-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYY
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | 分子名称: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, HYPOTHETICAL PROTEIN YDCE | | 著者 | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | 登録日 | 2002-04-30 | | 公開日 | 2002-10-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1H8Y
 
 | | Crystal structure of the class D beta-lactamase OXA-13 in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BETA-LACTAMASE, SULFATE ION | | 著者 | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | 登録日 | 2001-02-17 | | 公開日 | 2001-07-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
