6A6T
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | 著者 | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | 登録日 | 2018-06-29 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | 分子名称: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | 著者 | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | 登録日 | 2000-05-07 | | 公開日 | 2000-06-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6A6V
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with 7 additional mutations, in complex with FSA | | 分子名称: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | 著者 | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | 登録日 | 2018-06-29 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | 登録日 | 2018-07-20 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.797 Å) | | 主引用文献 | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABR
 
 | | Actin interacting protein 5 (Aip5, wild type) | | 分子名称: | Actin binding protein | | 著者 | Sun, J, Xie, Y, Toh, J.D.W, Hong, W, MIao, Y, Gao, Y.G. | | 登録日 | 2018-07-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
1EZW
 
 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | 分子名称: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | 著者 | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | 登録日 | 2000-05-12 | | 公開日 | 2000-09-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F07
 
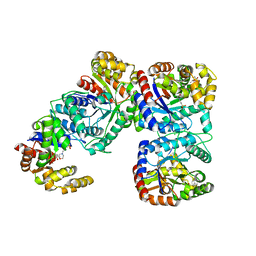 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | 著者 | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | 登録日 | 2000-05-15 | | 公開日 | 2000-09-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
6A82
 
 | |
6A6R
 
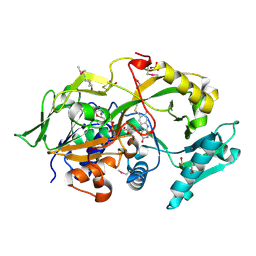 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | 著者 | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | 登録日 | 2018-06-29 | | 公開日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.609 Å) | | 主引用文献 | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6AE6
 
 | |
6AGF
 
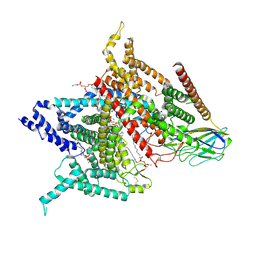 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | 登録日 | 2018-08-11 | | 公開日 | 2018-10-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
6ACC
 
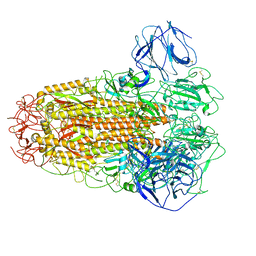 | |
6ACD
 
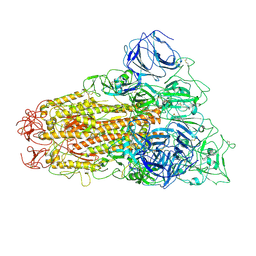 | |
1F4P
 
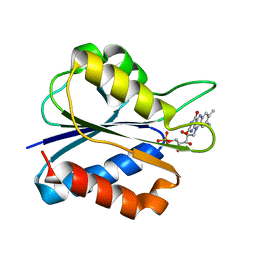 | |
1EZI
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | 分子名称: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE | | 著者 | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | 登録日 | 2000-05-11 | | 公開日 | 2001-02-14 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
1EZR
 
 | | CRYSTAL STRUCTURE OF NUCLEOSIDE HYDROLASE FROM LEISHMANIA MAJOR | | 分子名称: | CALCIUM ION, NUCLEOSIDE HYDROLASE | | 著者 | Shi, W, Schramm, V.L, Almo, S.C. | | 登録日 | 2000-05-11 | | 公開日 | 2000-05-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure.
J.Biol.Chem., 274, 1999
|
|
6AE7
 
 | |
1E2A
 
 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | 分子名称: | ENZYME IIA, MAGNESIUM ION | | 著者 | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | 登録日 | 1997-04-25 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
6AE5
 
 | |
6ALZ
 
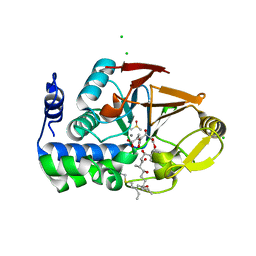 | | Crystal structure of Protein Phosphatase 1 bound to the natural inhibitor Tautomycetin | | 分子名称: | (2Z)-2-[(1R)-3-{[(2R,3S,4R,7S,8S,11S,13R,16E)-17-ethyl-4,8-dihydroxy-3,7,11,13-tetramethyl-6,15-dioxononadeca-16,18-dien-2-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2017-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | PP1:Tautomycetin Complex Reveals a Path toward the Development of PP1-Specific Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5ZV6
 
 | |
1FQJ
 
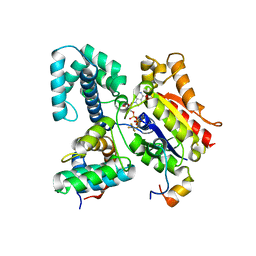 | | CRYSTAL STRUCTURE OF THE HETEROTRIMERIC COMPLEX OF THE RGS DOMAIN OF RGS9, THE GAMMA SUBUNIT OF PHOSPHODIESTERASE AND THE GT/I1 CHIMERA ALPHA SUBUNIT [(RGS9)-(PDEGAMMA)-(GT/I1ALPHA)-(GDP)-(ALF4-)-(MG2+)] | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(t) subunit alpha-1,Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(t) subunit alpha-1, MAGNESIUM ION, ... | | 著者 | Slep, K.C, Kercher, M.A, He, W, Cowan, C.W, Wensel, T.G, Sigler, P.B. | | 登録日 | 2000-09-05 | | 公開日 | 2001-02-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A.
Nature, 409, 2001
|
|
1FRY
 
 | | THE SOLUTION STRUCTURE OF SHEEP MYELOID ANTIMICROBIAL PEPTIDE, RESIDUES 1-29 (SMAP29) | | 分子名称: | MYELOID ANTIMICROBIAL PEPTIDE | | 著者 | Tack, B.F, Sawai, M.V, Kearney, W.R, Robertson, A.D, Sherman, M.A, Wang, W, Hong, T, Boo, L.M, Wu, H, Waring, A.J, Lehrer, R.I. | | 登録日 | 2000-09-07 | | 公開日 | 2002-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SMAP-29 has two LPS-binding sites and a central hinge.
Eur.J.Biochem., 269, 2002
|
|
1FE1
 
 | | CRYSTAL STRUCTURE PHOTOSYSTEM II | | 分子名称: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, CADMIUM ION, CHLOROPHYLL A, ... | | 著者 | Zouni, A, Witt, H.-T, Kern, J, Fromme, P, Krauss, N, Saenger, W, Orth, P. | | 登録日 | 2000-07-20 | | 公開日 | 2001-02-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution.
Nature, 409, 2001
|
|
1FKC
 
 | | HUMAN PRION PROTEIN (MUTANT E200K) FRAGMENT 90-231 | | 分子名称: | PRION PROTEIN | | 著者 | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | 登録日 | 2000-08-09 | | 公開日 | 2000-09-21 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
