5FTE
 
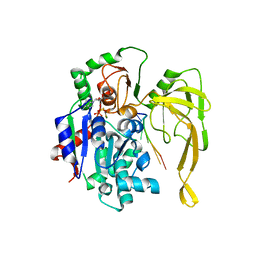 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | 分子名称: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | 著者 | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | 登録日 | 2016-01-12 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
8PCB
 
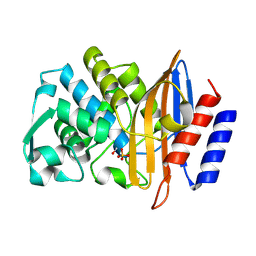 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 80 ms | | 分子名称: | BORATE ION, Beta-lactamase, SULFATE ION | | 著者 | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | 登録日 | 2023-06-11 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Time resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification
To Be Published
|
|
8PCO
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 150 ms | | 分子名称: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | 著者 | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | 登録日 | 2023-06-11 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Time resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification
To Be Published
|
|
5FTC
 
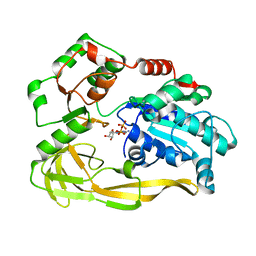 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, TPR DOMAIN PROTEIN | | 著者 | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | 登録日 | 2016-01-12 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.269 Å) | | 主引用文献 | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FV4
 
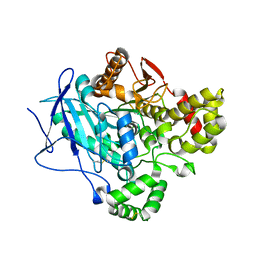 | |
5FHF
 
 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | 著者 | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | 登録日 | 2015-12-22 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | 分子名称: | CHLORIDE ION, UL31, UL34 protein, ... | | 著者 | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | 登録日 | 2015-10-16 | | 公開日 | 2016-03-16 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (35 Å) | | 主引用文献 | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
5FTD
 
 | | Crystal structure of Pif1 helicase from Bacteroides apo form | | 分子名称: | PHOSPHATE ION, TPR DOMAIN PROTEIN | | 著者 | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | 登録日 | 2016-01-12 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
8I7L
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with a novel inhibitor | | 分子名称: | 1-[3-[(4-chloranyl-2-fluoranyl-phenyl)carbamoylamino]-4-[cyclohexyl(2-methylpropyl)amino]phenyl]pyrrole-2-carboxylic acid, Indoleamine 2,3-dioxygenase 1, THIOSULFATE | | 著者 | Li, K, Liu, W, Dong, X. | | 登録日 | 2023-02-01 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Apo-Form Selective Inhibition of IDO for Tumor Immunotherapy.
J Immunol., 209, 2022
|
|
5FFP
 
 | | Crystal structure of CdiI from Burkholderia dolosa AUO158 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Immunity 23 family protein | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2015-12-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of CdiI from Burkholderia dolosa AUO158
To Be Published
|
|
5FHE
 
 | | Crystal structure of Bacteroides Pif1 bound to ssDNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | 著者 | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | 登録日 | 2015-12-22 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
1SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
5FGL
 
 | | Co-crystal Structure of NicR2_Hsp | | 分子名称: | 4-oxidanylidene-4-(6-oxidanylidene-1~{H}-pyridin-3-yl)butanoic acid, NicR | | 著者 | Zhang, K, Tang, H, Wu, G, Wang, W, Hu, H, Xu, P. | | 登録日 | 2015-12-21 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Co-crystal Structure of NicR2_Hsp
To Be Published
|
|
1SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
5FHH
 
 | | Structure of human Pif1 helicase domain residues 200-641 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, TETRAFLUOROALUMINATE ION | | 著者 | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | 登録日 | 2015-12-22 | | 公開日 | 2016-03-30 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FRD
 
 | | Structure of a thermophilic esterase | | 分子名称: | CARBOXYLESTERASE (EST-2), CHLORIDE ION, CITRATE ANION, ... | | 著者 | Sayer, C, Finnigan, W, Isupov, M.N, Levisson, M, Kengen, S.W.M, van der Oost, J, Harmer, N, Littlechild, J.A. | | 登録日 | 2015-12-17 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and Biochemical Characterisation of Archaeoglobus Fulgidus Esterase Reveals a Bound Coa Molecule in the Vicinity of the Active Site.
Sci.Rep., 6, 2016
|
|
8QAV
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN and IG2 | | 分子名称: | (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | 著者 | Witek, W, Ruszkowski, M. | | 登録日 | 2023-08-23 | | 公開日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.23 Å) | | 主引用文献 | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
1RS9
 
 | | Bovine endothelial NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | 著者 | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | 登録日 | 2003-12-09 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
8QAY
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, ACT, CIT, EDO, SO4 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CITRIC ACID, ... | | 著者 | Witek, W, Ruszkowski, M. | | 登録日 | 2023-08-23 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAW
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, IMD, EDO, FMT, GOL and TRS | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Witek, W, Ruszkowski, M. | | 登録日 | 2023-08-23 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
5EGN
 
 | |
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | 分子名称: | Nucleoprotein | | 著者 | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | 登録日 | 2015-11-12 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | 分子名称: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-12 | | 公開日 | 2015-12-23 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
8IDE
 
 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | 分子名称: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | 著者 | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | 登録日 | 2023-02-13 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
2OBI
 
 | |
