8E3N
 
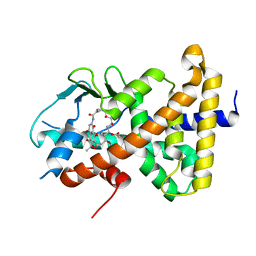 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | 分子名称: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | 著者 | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | 登録日 | 2022-08-17 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2BNZ
 
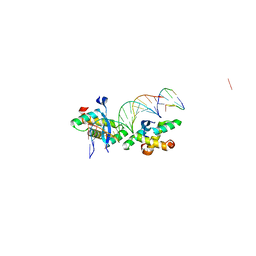 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to inverted DNA heptad repeats | | 分子名称: | 5'-D(*CP*TP*AP*AP*TP*CP*AP*CP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*GP *TP*GP*AP*TP*TP*AP*GP*C)-3', ORF OMEGA | | 著者 | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | 登録日 | 2005-04-06 | | 公開日 | 2006-03-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
8EBU
 
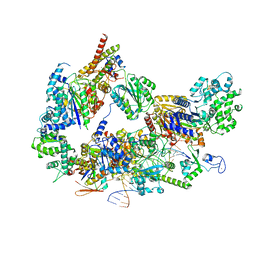 | | XPC release from Core7-XPA-DNA (Cy5) | | 分子名称: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | 著者 | Kim, J, Yang, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBS
 
 | |
8EBT
 
 | |
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | 分子名称: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | 著者 | Kim, J, Yang, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBV
 
 | |
8EBW
 
 | |
8EBX
 
 | |
3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | 分子名称: | Tumor necrosis factor receptor superfamily member 21 | | 著者 | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | 登録日 | 2011-10-06 | | 公開日 | 2012-05-02 | | 最終更新日 | 2012-07-11 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8E3S
 
 | |
2C1Z
 
 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | 分子名称: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | 著者 | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | 登録日 | 2005-09-22 | | 公開日 | 2006-01-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2C1X
 
 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | 著者 | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | 登録日 | 2005-09-22 | | 公開日 | 2006-01-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
8EC0
 
 | | III2IV respiratory supercomplex from Saccharomyces cerevisiae cardiolipin-lacking mutant | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | 著者 | Hryc, C.F, Mileykovskaya, E, Baker, M, Dowhan, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex.
Nat Commun, 14, 2023
|
|
3U2Z
 
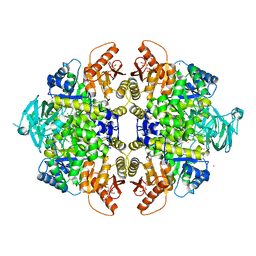 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | 著者 | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-10-04 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
8E7S
 
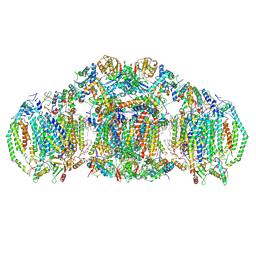 | | III2IV2 respiratory supercomplex from Saccharomyces cerevisiae with 4 bound UQ6 | | 分子名称: | (5S,11R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-4,6,10,12,16-pentaoxa-5,11-diphosphaoctadec-1-yl pentadecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | 著者 | Hryc, C.F, Mileykovskaya, E, Baker, M, Dowhan, W. | | 登録日 | 2022-08-24 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex.
Nat Commun, 14, 2023
|
|
2DIS
 
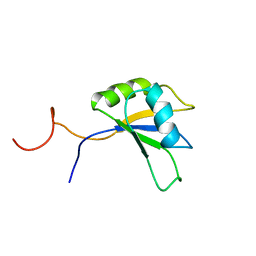 | | Solution structure of the RRM domain of unnamed protein product | | 分子名称: | unnamed protein product | | 著者 | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-30 | | 公開日 | 2006-09-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RRM domain of unnamed protein product
To be Published
|
|
8A2T
 
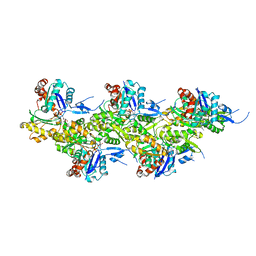 | | Cryo-EM structure of F-actin in the Mg2+-ADP nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.24 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2Z
 
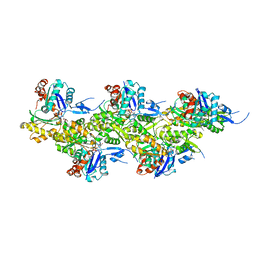 | | Cryo-EM structure of F-actin in the Ca2+-ADP nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.15 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2U
 
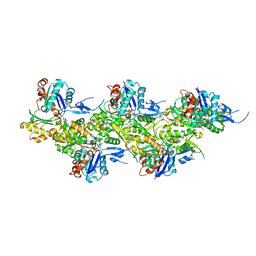 | | Cryo-EM structure of F-actin in the Ca2+-ADP-BeF3- nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2Y
 
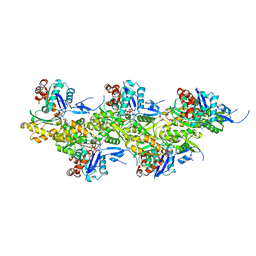 | | Cryo-EM structure of F-actin in the Ca2+-ADP-Pi nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.15 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2S
 
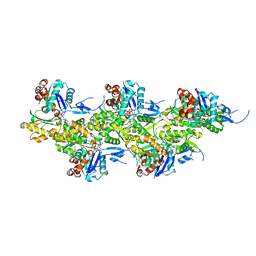 | | Cryo-EM structure of F-actin in the Mg2+-ADP-Pi nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.22 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2R
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP-BeF3- nucleotide state. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | 登録日 | 2022-06-06 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.17 Å) | | 主引用文献 | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
2DIW
 
 | | Solution structure of the RPR domain of Putative RNA-binding protein 16 | | 分子名称: | Putative RNA-binding protein 16 | | 著者 | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-30 | | 公開日 | 2006-09-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RPR domain of Putative RNA-binding protein 16
To be Published
|
|
2DEA
 
 | |
