5A4N
 
 | |
5A7T
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
7CRP
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
5A62
 
 | | Hydrolytic potential of the ammonia-oxidizing Thaumarchaeon Nitrososphaera gargenis - crystal structure and activity profiles of carboxylesterases linked to their metabolic function | | Descriptor: | ACETATE ION, PUTATIVE ALPHA/BETA HYDROLASE FOLD PROTEIN | | Authors: | Chow, J, Kaljunen, H, Nittinger, E, Spieck, E, Rarey, M, Mueller-Dieckmann, J, Streit, W.R. | | Deposit date: | 2015-06-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrolytic Potential of the Ammonia-Oxidizing Thaumarchaeon Nitrososphaera Gargenis - Crystal Structure and Activity Profiles of Carboxylesterases Linked to Their Metabolic Function
To be Published
|
|
7MCO
 
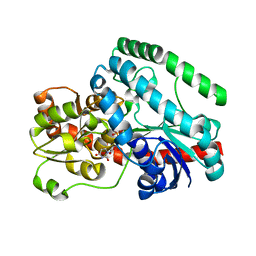 | | Crystal Structure of Tetur04g02350 | | Descriptor: | UDP-glycosyltransferase 203A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Danehsian, L, Kluza, A, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Tetur04g02350
To Be Published
|
|
7MPF
 
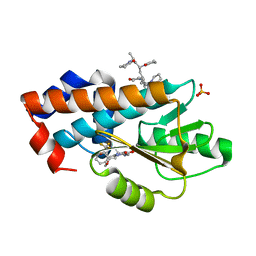 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5A63
 
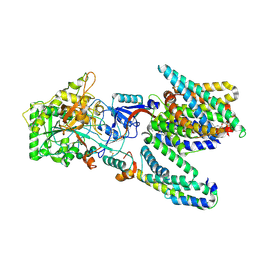 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
4ZTZ
 
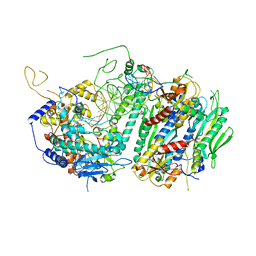 | |
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MRP
 
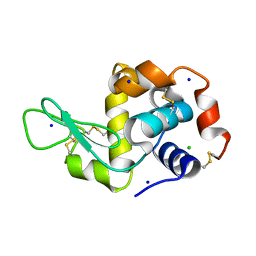 | |
7MTY
 
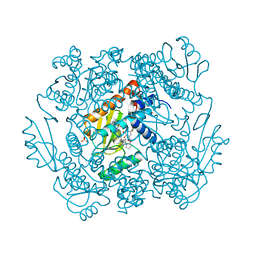 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988569 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
4ZRV
 
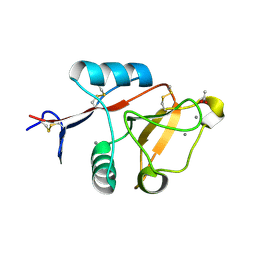 | | Structure of cow mincle CRD complexed with trehalose mono butyrate | | Descriptor: | 6-O-butanoyl-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Feinberg, H, Rambaruth, N.D.S, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
7MVZ
 
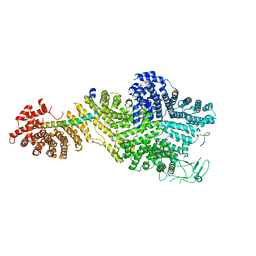 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96-Nup145N complex (Nup188 residues 1-1858; Nic96 residues 240-301; Nup145N residues 640-732) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5AGF
 
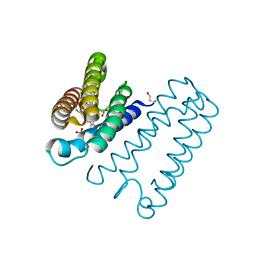 | | Nitrosyl complex of the D121Q variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C PRIME, HEME C, NITRIC OXIDE, ... | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2015-01-30 | | Release date: | 2015-09-09 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
4ZWT
 
 | |
5ABG
 
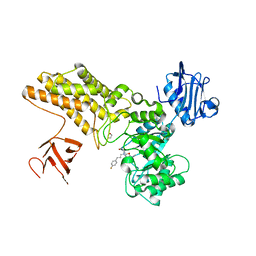 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AJ8
 
 | |
4ZSC
 
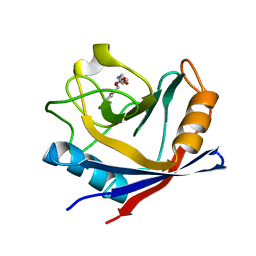 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
4ZSH
 
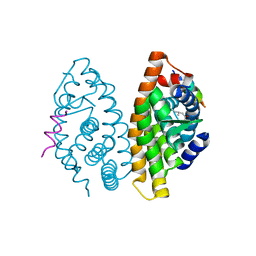 | | RXR LBD in complex with 9-cis-13,14-dihydroretinoic acid | | Descriptor: | (5S,6S,9R,13R)-2,3-didehydro-5,6,7,8,9,10,11,12,13,14-decahydroretinoic acid, NCoA2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Rochel, N, Krezel, W, Ruhl, R. | | Deposit date: | 2015-05-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice.
Plos Genet., 11, 2015
|
|
7CRR
 
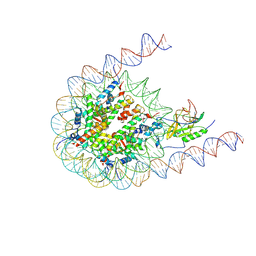 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
5AEM
 
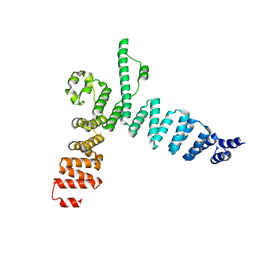 | | Structure of t131 N-terminal TPR array | | Descriptor: | TRANSCRIPTION FACTOR TAU 131 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Muller, C.W. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly.
Nat Commun, 6, 2015
|
|
7CV0
 
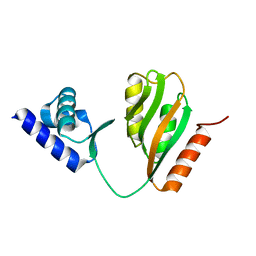 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
4ZRS
 
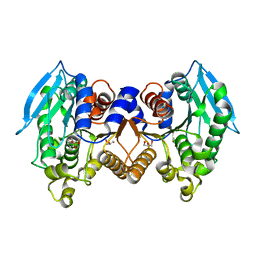 | | Crystal structure of a cloned feruloyl esterase from a soil metagenomic library | | Descriptor: | Esterase, GLYCEROL | | Authors: | Xie, W, Chen, R, Cao, L, Liu, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing the Thermostability of Feruloyl Esterase EstF27 by Directed Evolution and the Underlying Structural Basis
J.Agric.Food Chem., 63, 2015
|
|
5A1W
 
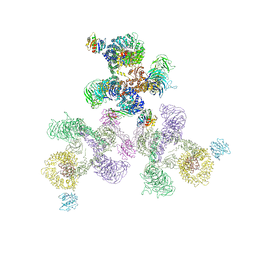 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
