7GQB
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z44548882 | | Descriptor: | 1,1-diphenylmethanamine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
6KM9
 
 | | Crystal structure of SucA from Vibrio vulnificus | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
7GQR
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z235343929 | | Descriptor: | 2-chloro-1,3-benzoxazole, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
6DPE
 
 | |
7GPO
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with POB0066 | | Descriptor: | (3R,4S)-1-acetyl-4-phenylpyrrolidine-3-carboxylic acid, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198152494 | | Descriptor: | 1H-imidazole-5-carbonitrile, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
8IQC
 
 | |
8IQI
 
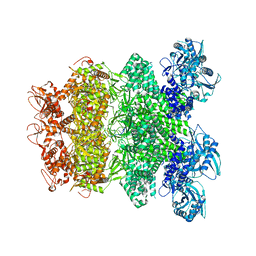 | | Structure of Full-Length AsfvPrimPol in Complex-Form | | Descriptor: | DNA (32-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Shao, Z.W, Su, S.C, Gan, J.H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures and implications of the C962R protein of African swine fever virus.
Nucleic Acids Res., 51, 2023
|
|
1DGN
 
 | | SOLUTION STRUCTURE OF ICEBERG, AN INHIBITOR OF INTERLEUKIN-1BETA GENERATION | | Descriptor: | ICEBERG (PROTEASE INHIBITOR) | | Authors: | Humke, E.W, Shriver, S.K, Starovasnik, M.A, Fairbrother, W.J, Dixit, V.M. | | Deposit date: | 1999-11-24 | | Release date: | 2000-10-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ICEBERG: a novel inhibitor of interleukin-1beta generation.
Cell(Cambridge,Mass.), 103, 2000
|
|
8IQH
 
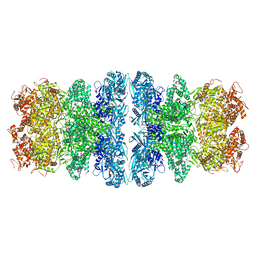 | |
6DUB
 
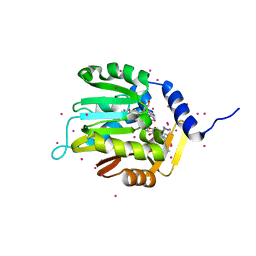 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
8IQB
 
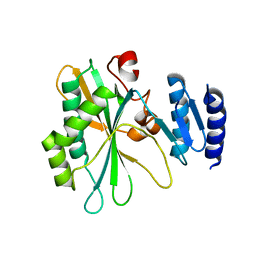 | |
6KHV
 
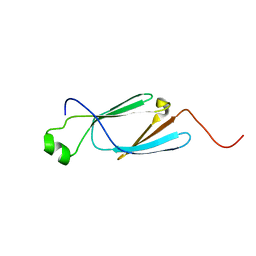 | | Solution Structure of the CS2 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
2VSH
 
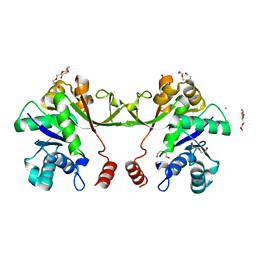 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
3NAE
 
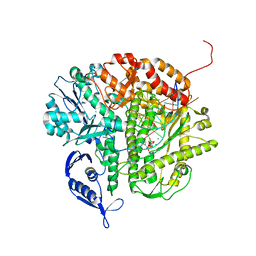 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite Guanidinohydantoin | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA Polymerase Allows dAMP and dGMP To Be Inserted opposite Guanidinohydantoin .
Biochemistry, 49, 2010
|
|
6KJU
 
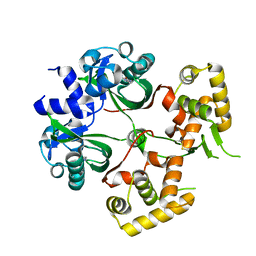 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
8J1F
 
 | | GSK101 bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1D
 
 | | Cryo-EM structure of apo state mTRPV4 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
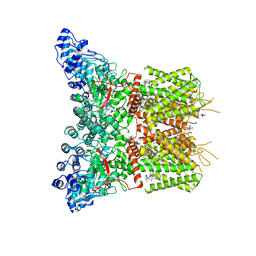 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8QMO
 
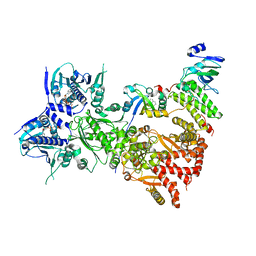 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
4AAU
 
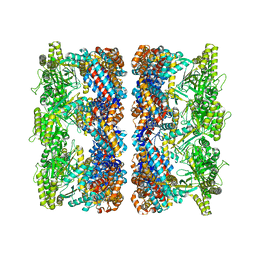 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
3RPM
 
 | |
4Q3S
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABHPE | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
1BPN
 
 | |
