821P
 
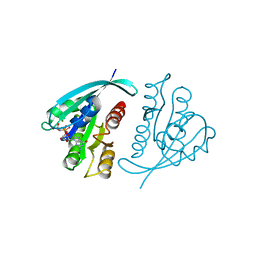 | | THREE-DIMENSIONAL STRUCTURES AND PROPERTIES OF A TRANSFORMING AND A NONTRANSFORMING GLYCINE-12 MUTANT OF P21H-RAS | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Scheidig, A.J, Krengel, U, Pai, E.F, Kabsch, W, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras.
Biochemistry, 32, 1993
|
|
2BMG
 
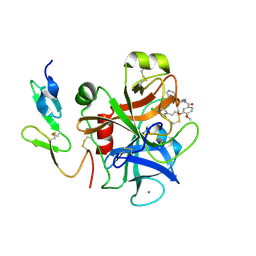 | | Crystal structure of factor Xa in complex with 50 | | Descriptor: | 3-[2-(2,4-DICHLOROPHENYL)ETHOXY]-4-METHOXY-N-[(1-PYRIDIN-4-YLPIPERIDIN-4-YL)METHYL]BENZAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Schreuder, H, Matter, H, Will, D.W, Nazare, M, Laux, V, Wehner, V, Loenze, P, Liesum, A. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Requirements for Factor Xa Inhibition by 3-Oxybenzamides with Neutral P1 Substituents: Combining X-Ray Crystallography, 3D-Qsar and Tailored Scoring Functions
J.Med.Chem., 48, 2005
|
|
2C1I
 
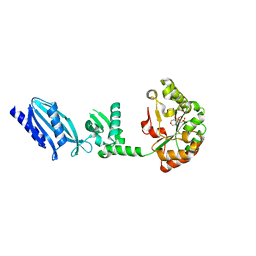 | | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) D 275 N Mutant. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PEPTIDOGLYCAN GLCNAC DEACETYLASE, SULFATE ION, ... | | Authors: | Blair, D.E, Schuttelkopf, A.W, MacRae, J.I, van Aalten, D.M.F. | | Deposit date: | 2005-09-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
2BUE
 
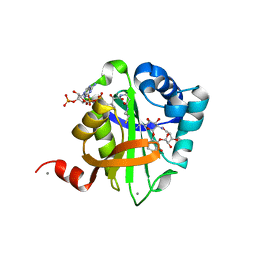 | | Structure of AAC(6')-Ib in complex with Ribostamycin and Coenzyme A. | | Descriptor: | AAC(6')-IB, CALCIUM ION, COENZYME A, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
4KT6
 
 | | High-resolution crystal structure Streptococcus pyogenes beta-NAD+ glycohydrolase in complex with its endogenous inhibitor IFS reveals a water-rich interface | | Descriptor: | Nicotine adenine dinucleotide glycohydrolase, Putative uncharacterized protein | | Authors: | Yoon, J.Y, An, D.R, Yoon, H.-J, Kim, H.S, Lee, S.J, Im, H.N, Jang, J.Y, Suh, S.W. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution crystal structure of Streptococcus pyogenes beta-NAD(+) glycohydrolase in complex with its endogenous inhibitor IFS reveals a highly water-rich interface
J.SYNCHROTRON RADIAT., 20, 2013
|
|
2C1F
 
 | |
2BT1
 
 | | Epstein Barr Virus dUTPase in complex with a,b-imino dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MAGNESIUM ION | | Authors: | Tarbouriech, N, Buisson, M, Seigneurin, J.-M, Cusack, S, Burmeister, W.P. | | Deposit date: | 2005-05-24 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Monomeric Dutpase from Epstein-Barr Virus Mimics Trimeric Dutpases
Structure, 13, 2005
|
|
1S3R
 
 | | Crystal structure of the human-specific toxin intermedilysin | | Descriptor: | SULFATE ION, intermedilysin | | Authors: | Polekhina, G, Giddings, K.S, Tweten, R.K, Parker, M.W. | | Deposit date: | 2004-01-14 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2TMP
 
 | | N-TERMINAL DOMAIN OF TISSUE INHIBITOR OF METALLOPROTEINASE-2 (N-TIMP-2), NMR, 49 STRUCTURES | | Descriptor: | TISSUE INHIBITOR OF METALLOPROTEINASES-2 | | Authors: | Muskett, F.W, Frenkiel, T.A, Feeney, J, Freedman, R.B, Carr, M.D, Williamson, R.A. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the N-terminal domain of tissue inhibitor of metalloproteinases-2 and characterization of its interaction site with matrix metalloproteinase-3.
J.Biol.Chem., 273, 1998
|
|
8A0J
 
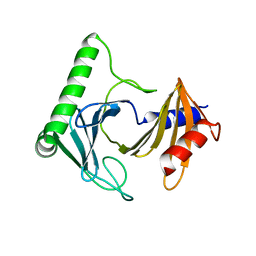 | | Crystal structure of the kinetoplastid kinetochore protein Trypanosoma congolense KKT2 divergent polo-box domain | | Descriptor: | Uncharacterized protein TCIL3000_11_11110 | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
5RVZ
 
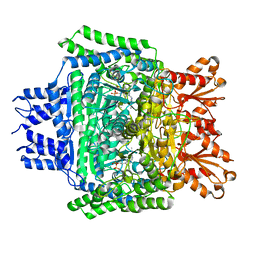 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z1929757385 | | Descriptor: | (3S)-1-(3-fluoropyridin-2-yl)-4,4-dimethylpyrrolidin-3-ol, MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1SGE
 
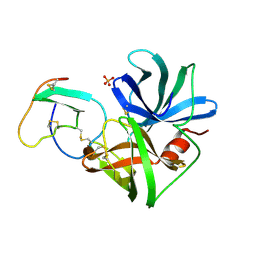 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2BKB
 
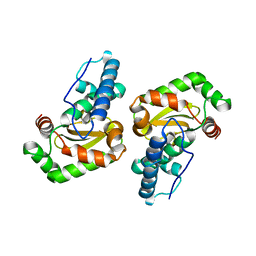 | | q69e-FeSOD | | Descriptor: | FE (II) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Yikilmaz, E, Rodgers, D.W, Miller, A.-F. | | Deposit date: | 2005-02-14 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crucial Importance of Chemistry in the Structure-Function Link: Manipulating Hydrogen Bonding in Iron-Containing Superoxide Dismutase.
Biochemistry, 45, 2006
|
|
2BUQ
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 in Complex with Catechol | | Descriptor: | CATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BWD
 
 | | Atomic Resolution Structure of Achromobacter cycloclastes Cu Nitrite Reductase with Endogenously bound Nitrite and NO | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C0D
 
 | | Structure of the mitochondrial 2-cys peroxiredoxin from Plasmodium falciparum | | Descriptor: | THIOREDOXIN PEROXIDASE 2 | | Authors: | Boucher, I.W, Brannigan, J.A, Wilkinson, A.J, Brzozowski, A.M, Muller, S. | | Deposit date: | 2005-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Biochemical Characterisation of a Mitochondrial Peroxiredoxin from Plasmodium Falciparum
Mol.Microbiol., 61, 2006
|
|
1SI6
 
 | |
2BUY
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with Catechol | | Descriptor: | CATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
6PS5
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
2BUT
 
 | | Crystal Structure Of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S - APO | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BYI
 
 | | 3-(5-chloro-2,4-dihydroxyphenyl)-pyrazole-4-carboxamides as Inhibitors of the Hsp90 Molecular Chaperone | | Descriptor: | 3-(5-CHLORO-2,4-DIHYDROXY-PHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID 4-SULFAMOYL-BENZYLAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Brough, P.A, Barril, X, Beswick, M, Dymock, B.W, Drysdale, M.J, Wright, L, Grant, K, Massey, A, Surgenor, A, Workman, P. | | Deposit date: | 2005-08-02 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 3-(5-Chloro-2,4-Dihydroxyphenyl)-Pyrazole-4-Carboxamides as Inhibitors of the Hsp90 Molecular Chaperone.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BV0
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with Protocatechuate. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
6QAF
 
 | | Crystal structure of human Arginase-1 at pH 9.0 in complex with CB-1158/INCB001158 | | Descriptor: | Arginase-1, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Tabak, W.W.A, Zaman, G.J.R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
2BUZ
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with 4-Nitrocatechol | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases
Ph D Thesis, 2001
|
|
2C0B
 
 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
