5A7Z
 
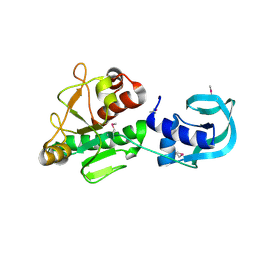 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.1 angstrom resolution. | | Descriptor: | TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
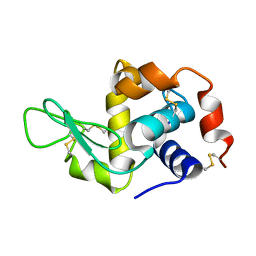
5A3E
 
 | |
5ABT
 
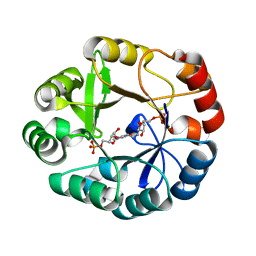 | | S.enterica HisA mutant D7N, G102A, V106M, D176A | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Protein Structures, Functions and Dynamics of Adaptive Evolution
To be Published
|
|
5A3K
 
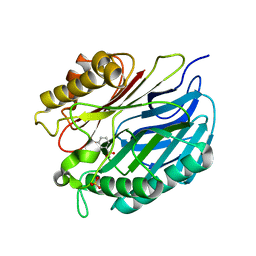 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUV
 
 | |
9BLG
 
 | | Crystal structure of non-receptor protein tyrosine phosphatase SHP2 in complex with PF-07284892 | | Descriptor: | (1S)-1'-{6-[(2-amino-3-chloropyridin-4-yl)sulfanyl]-1,2,4-triazin-3-yl}-1,3-dihydrospiro[indene-2,4'-piperidin]-1-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | SHP2 Inhibition Sensitizes Diverse Oncogene-Addicted Solid Tumors to Re-treatment with Targeted Therapy.
Cancer Discov, 13, 2023
|
|
8YVV
 
 | | The Crystal Structure of BTK from Biortus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of BTK from Biortus.
To Be Published
|
|
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8XRP
 
 | | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 receptor subunit beta-2, ... | | Authors: | Chen, H.Q, Wang, X.Q, Ge, X.F, Zeng, J.W, Wang, J.W. | | Deposit date: | 2024-01-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly of the human IL-12 signaling complex.
Structure, 2024
|
|
8YSA
 
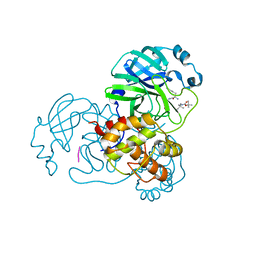 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H102 | | Descriptor: | 3C-like proteinase nsp5, BOC-TBG-PHE-ELL | | Authors: | Zheng, W.Y, Fu, L.F, Feng, Y, Han, P, Qi, J.X. | | Deposit date: | 2024-03-22 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery, Biological Activity, and Structural Mechanism of a Potent Inhibitor of SARS-CoV-2 Main Protease
To Be Published
|
|
8ZGD
 
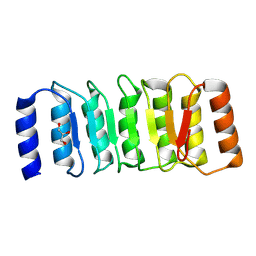 | | The Crystal Structure of the NLRP1_LRR domain from Biortus. | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-09 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the NLRP1_LRR domain from Biortus.
To Be Published
|
|
6XE0
 
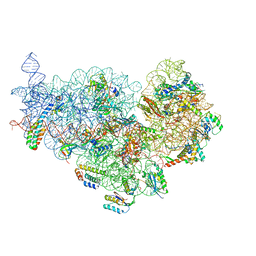 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
8ZKD
 
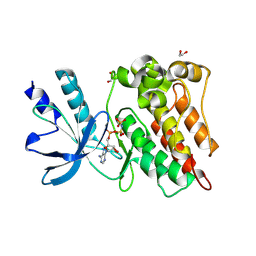 | | The Crystal Structure of the RON from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Macrophage-stimulating protein receptor beta chain, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the RON from Biortus.
To Be Published
|
|
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
4W8Q
 
 | |
4WED
 
 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | Descriptor: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | Authors: | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
4V9I
 
 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4W4S
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum in complex with BPH-629 | | Descriptor: | Uncharacterized protein blr2150, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4W89
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
