8CH7
 
 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CGF
 
 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6E3Y
 
 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
4UUH
 
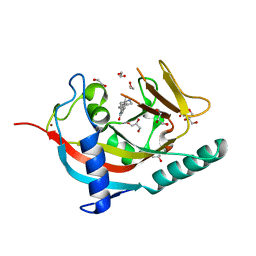 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-3-[4-(piperazin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | Deposit date: | 2014-07-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1- Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
8CAQ
 
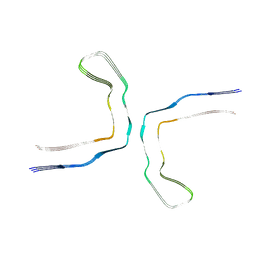 | | Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis | | Descriptor: | Microtubule-associated protein tau | | Authors: | Qi, C, Hasegawa, M, Takao, M, Sakai, M, Akagi, M, Iwasaki, Y, Yoshida, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Identical tau filaments in subacute sclerosing panencephalitis and chronic traumatic encephalopathy.
Acta Neuropathol Commun, 11, 2023
|
|
4UWO
 
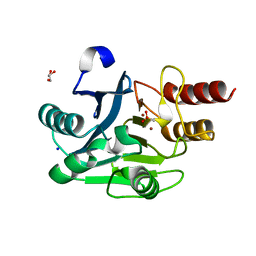 | | Native di-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4UWR
 
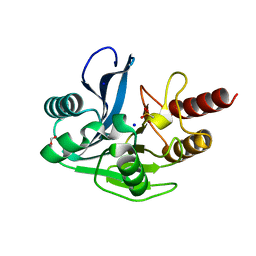 | | Mono-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | FORMIC ACID, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
6E52
 
 | |
4V07
 
 | | Dimeric pseudorabies virus protease pUL26N at 2.1 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, UL26 | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
6E28
 
 | | The CARD9 CARD domain-swapped dimer | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Ferrao, R, Boenig, G, Deuber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Picomolar zinc binding modulates formation of Bcl10-nucleating assemblies of the caspase recruitment domain (CARD) of CARD9.
J. Biol. Chem., 293, 2018
|
|
4UY3
 
 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4UXV
 
 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
6EBS
 
 | | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, R.A.G, Pinheiro, M.P, de Souza, A.L, Hunter, W.N, Nonato, M.C. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate
To Be Published
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
6EHM
 
 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6E31
 
 | | Crystal structure of RIAM in an autoinhibited configuration. | | Descriptor: | Rap1-interacting adapter molecule | | Authors: | Chang, Y.C, Su, W, Zhang, H, Huang, Q, Philips, M.R, Wu, J. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for autoinhibition of RIAM regulated by FAK in integrin activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EIK
 
 | | A de novo designed heptameric coiled coil CC-Hept-I24E | | Descriptor: | CC-Hept-I24E, FORMIC ACID, GLYCEROL | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
4UYI
 
 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
6E5J
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, beta3 helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, beta3 helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
4V35
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CALCIUM ION, ... | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6E5Y
 
 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
