4EXM
 
 | | The crystal structure of an engineered phage lysin containing the binding domain of pesticin and the killing domain of T4-lysozyme | | 分子名称: | Pesticin, Lysozyme Chimera | | 著者 | Seddiki, N, Noinaj, N, Fairman, J.W, Lukacik, P, Barnard, T.J, Buchanan, S.K. | | 登録日 | 2012-04-30 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6N9V
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS1) | | 分子名称: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Fox, T, Val, N, Yang, W. | | 登録日 | 2018-12-04 | | 公開日 | 2019-03-06 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
5VA3
 
 | |
5VA2
 
 | |
6MIT
 
 | | LptBFGC from Enterobacter cloacae | | 分子名称: | CHLORIDE ION, LPS export ABC transporter permease LptF, Lipopolysaccharide export system ATP-binding protein, ... | | 著者 | Owens, T.W, Kahne, D, Kruse, A.C. | | 登録日 | 2018-09-20 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
2B9S
 
 | |
1ZJ5
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | 分子名称: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | 著者 | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | 登録日 | 2005-04-28 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZEQ
 
 | | 1.5 A Structure of apo-CusF residues 6-88 from Escherichia coli | | 分子名称: | Cation efflux system protein cusF | | 著者 | Loftin, I.R, Franke, S, Roberts, S.A, Weichsel, A, Heroux, A, Montfort, W.R, Rensing, C, McEvoy, M.M. | | 登録日 | 2005-04-19 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Novel Copper-Binding Fold for the Periplasmic Copper Resistance Protein CusF.
Biochemistry, 44, 2005
|
|
1ZIC
 
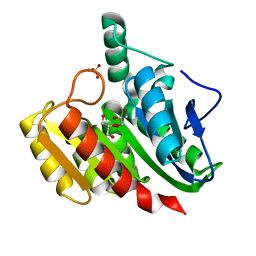 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S, R206A) mutant- 1.7 A | | 分子名称: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | 著者 | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | 登録日 | 2005-04-27 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4JRM
 
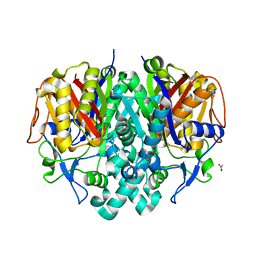 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio Cholerae (space group P212121) at 1.75 Angstrom | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, ACETATE ION, GLYCEROL | | 著者 | Hou, J, Chruszcz, M, Shabalin, I.G, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-03-21 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio cholerae (space group P43) at 2.2 Angstrom
To be Published
|
|
3TJI
 
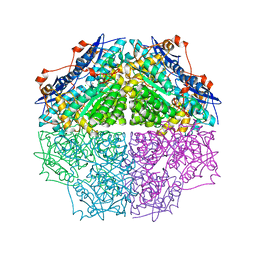 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ENTEROBACTER sp. 638 (EFI TARGET EFI-501662) with BOUND MG | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-08-24 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of an enolase from enterobacter sp. 638 (efi target efi-501662) with boung mg
to be published
|
|
5L7L
 
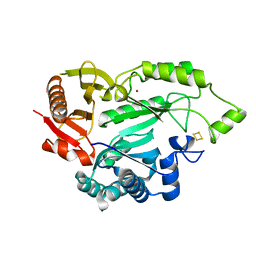 | |
3TL2
 
 | | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation. | | 分子名称: | 1,2-ETHANEDIOL, Malate dehydrogenase, THIOCYANATE ION | | 著者 | Blus, B.J, Chruszcz, M, Tkaczuk, K.L, Osinski, T, Cymborowski, M, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-29 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation.
TO BE PUBLISHED
|
|
5S4J
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | 分子名称: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.124 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1Y79
 
 | | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp in Complex with a Peptidic Inhibitor | | 分子名称: | ASPARTIC ACID, GLYCINE, LYSINE, ... | | 著者 | Comellas-Bigler, M, Lang, R, Bode, W, Maskos, K. | | 登録日 | 2004-12-08 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp: Further Indication of a Ligand-dependant Hinge Movement Mechanism
J.Mol.Biol., 349, 2005
|
|
5MUE
 
 | | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier | | 分子名称: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, Alpha-tocopherol transfer protein, CHLORIDE ION, ... | | 著者 | Stocker, A, Aeschimann, W. | | 登録日 | 2017-01-13 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier.
Sci Rep, 7, 2017
|
|
4J5F
 
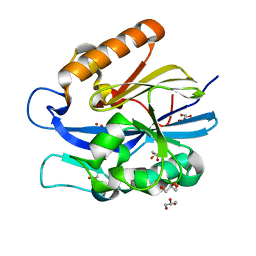 | | Crystal Structure of B. thuringiensis AiiA mutant F107W | | 分子名称: | GLYCEROL, N-acyl homoserine lactonase, ZINC ION | | 著者 | Liu, C.F, Liu, D, Momb, J, Thomas, P.W, Lajoie, A, Petsko, G.A, Fast, W, Ringe, D. | | 登録日 | 2013-02-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-gamma-lactonase from Bacillus thuringiensis.
Biochemistry, 52, 2013
|
|
7M4X
 
 | |
7M4Y
 
 | |
2JIZ
 
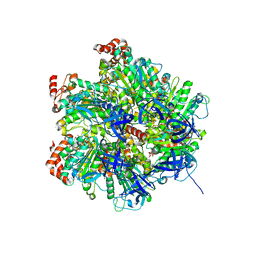 | | The Structure of F1-ATPase inhibited by resveratrol. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | 著者 | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | 登録日 | 2007-07-03 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5S4I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | 分子名称: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3OM7
 
 | | Crystal structure of B. megaterium levansucrase mutant Y247W | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Levansucrase, ... | | 著者 | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | 登録日 | 2010-08-26 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
7VPP
 
 | | Structures of a deltacoronavirus spike protein bound to porcine and human receptors indicate the risk of virus adaptation to humans | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ji, W, Xu, Y, Zhang, S. | | 登録日 | 2021-10-17 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structures of a deltacoronavirus spike protein bound to porcine and human receptors.
Nat Commun, 13, 2022
|
|
4A6N
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH TIGECYCLINE | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | 登録日 | 2011-11-07 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5V0P
 
 | | Crystal Structure of Beta-ketoacyl-ACP synthase III-2 (FabH2) (C113A) from Vibrio Cholerae co-crystallized with octanoyl-CoA | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, OCTANOYL-COENZYME A, SODIUM ION | | 著者 | Hou, J, Zheng, H, Cooper, D.R, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-28 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structure of Beta-ketoacyl-ACP synthase III-2 (FabH2) (C113A) from Vibrio Cholerae co-crystallized with octanoyl-CoA
To Be Published
|
|
