1GT5
 
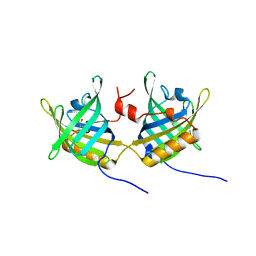 | | Complexe of Bovine Odorant Binding Protein with benzophenone | | Descriptor: | DIPHENYLMETHANONE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
2X32
 
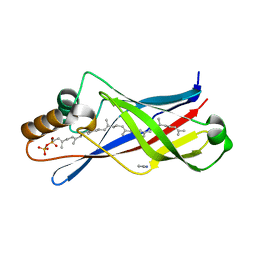 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, CELLULOSE-BINDING PROTEIN, IMIDAZOLE | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2X34
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | CELLULOSE-BINDING PROTEIN, X158, Ubiquinone-8 | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2XBZ
 
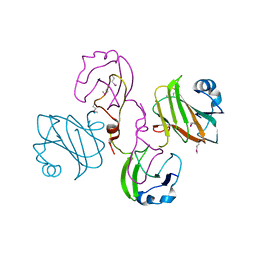 | | Multiple oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand-binding site | | Descriptor: | RETS-HYBRID SENSOR KINASE | | Authors: | Vincent, F, Round, A, Reynaud, A, Bordi, C, Filloux, A, Bourne, Y. | | Deposit date: | 2010-04-15 | | Release date: | 2010-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct Oligomeric Forms of the Pseudomonas Aeruginosa Rets Sensor Domain Modulate Accessibility to the Ligand Binding Site.
Environ.Microbiol., 12, 2010
|
|
2BKX
 
 | |
2BKV
 
 | |
2VHL
 
 | | The Three-dimensional structure of the N-Acetylglucosamine-6- phosphate deacetylase from Bacillus subtilis | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, FE (III) ION, N-ACETYLGLUCOSAMINE-6-PHOSPHATE DEACETYLASE, ... | | Authors: | Vincent, F, Yates, D, Garman, E, Davies, G.J. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Three-Dimensional Structure of the N-Acetylglucosamine-6-Phosphate Deacetylase from Bacillus Subtilis
J.Biol.Chem., 279, 2004
|
|
1UZ4
 
 | | Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, GLYCEROL, MAN5A, ... | | Authors: | Vincent, F, Davies, G.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Common Inhibition of Both Beta-Glucosidases and Beta-Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
1GT4
 
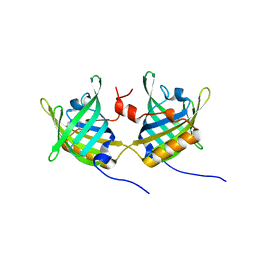 | | Complex of Bovine Odorant Binding Protein with undecanal | | Descriptor: | ODORANT-BINDING PROTEIN, UNDECANAL | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1GT3
 
 | | Complex of Bovine Odorant Binding Protein with dihydromyrcenol | | Descriptor: | (3S)-1-octen-3-ol, 2,6-DIMETHYL-7-OCTEN-2-OL, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1GT1
 
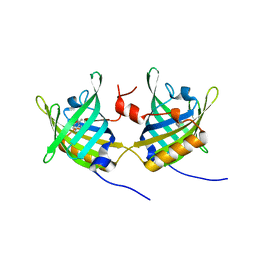 | | Complex of Bovine Odorant Binding Protein with Aminoanthracene and pyrazine | | Descriptor: | (3S)-1-octen-3-ol, 2-ISOBUTYL-3-METHOXYPYRAZINE, ANTHRACEN-1-YLAMINE, ... | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1G85
 
 | | CRYSTAL STRUCTURE OF BOVINE ODORANT BINDING PROTEIN COMPLEXED WITH IS NATURAL LIGAND | | Descriptor: | (3R)-oct-1-en-3-ol, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1E5P
 
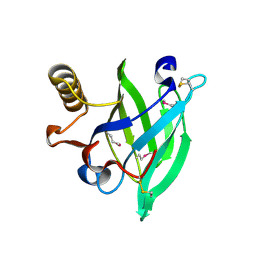 | | Crystal structure of aphrodisin, a sex pheromone from female hamster | | Descriptor: | APHRODISIN | | Authors: | Vincent, F, Brown, K, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of aphrodisin, a sex pheromone from female hamster.
J.Mol.Biol., 305, 2001
|
|
1DZP
 
 | | Porcine Odorant Binding Protein Complexed with diphenylmethanone | | Descriptor: | DIPHENYLMETHANONE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-12-06 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1DZM
 
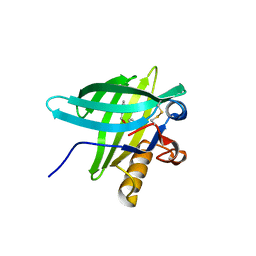 | | Porcine Odorant Binding Protein Complexed with benzoic acid phenylmethylester | | Descriptor: | BENZOIC ACID PHENYLMETHYLESTER, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Ramoni, R, Grolli, S, Pelosi, P, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1E02
 
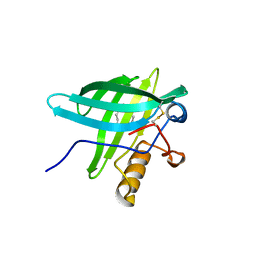 | | Porcine Odorant Binding Protein Complexed with undecanal | | Descriptor: | ODORANT-BINDING PROTEIN, UNDECANAL | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-09 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1E00
 
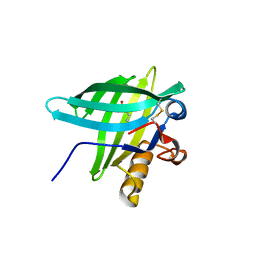 | | Porcine Odorant Binding Protein Complexed with 2,6-dimethyl-7-octen-2-ol | | Descriptor: | 2,6-DIMETHYL-7-OCTEN-2-OL, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-07 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1ODS
 
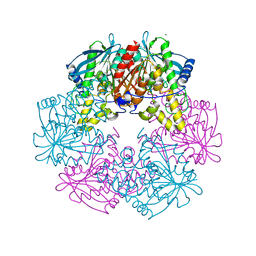 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1DZK
 
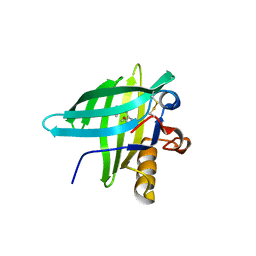 | | Porcine Odorant Binding Protein Complexed with pyrazine (2-isobutyl-3-metoxypyrazine) | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1E06
 
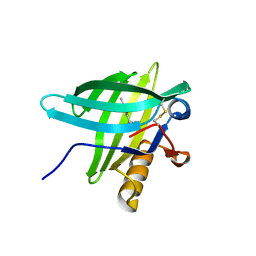 | | Porcine Odorant Binding Protein Complexed with 5-methyl-2-(1-methylethyl)phenol | | Descriptor: | 5-METHYL-2-(1-METHYLETHYL)PHENOL, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-10 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1DZJ
 
 | | Porcine Odorant Binding Protein Complexed with 2-amino-4-butyl-5-propylselenazole | | Descriptor: | 4-butyl-5-propyl-1,3-selenazol-2-amine, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1ODT
 
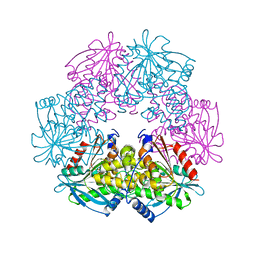 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
5O7J
 
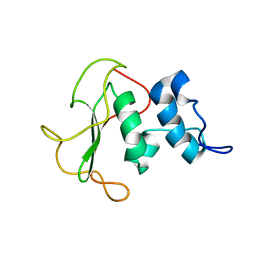 | | Structural insights into the periplasmic sensor domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa | | Descriptor: | Histidine kinase | | Authors: | Ali-Ahmad, A, Bornet, O, Fadel, F, Bourne, Y, Vincent, F, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa.
Sci Rep, 7, 2017
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
