1EWZ
 
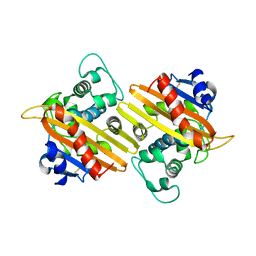 | | CRYSTAL STRUCTURE OF THE OXA-10 BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | BETA LACTAMASE OXA-10 | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Tranier, S, Ishiwata, A, Kotra, L.P, Samama, J.P, Mobashery, S. | | Deposit date: | 2000-04-28 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Structural and Mechanistic Insights for Class D beta-Lactamases: Evidence for a Novel Catalytic Process for Turnover of beta-Lactam Antibiotics
J.Am.Chem.Soc., 122, 2000
|
|
1K55
 
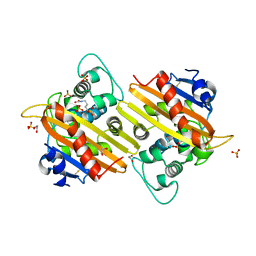 | | OXA 10 class D beta-lactamase at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Beta lactamase OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K57
 
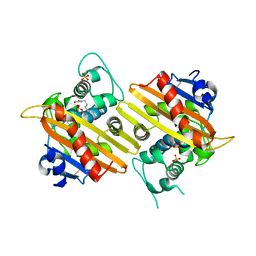 | | OXA 10 class D beta-lactamase at pH 6.0 | | Descriptor: | BETA LACTAMASE OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K56
 
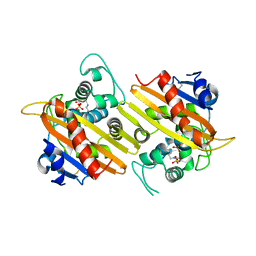 | | OXA 10 class D beta-lactamase at pH 6.5 | | Descriptor: | OXA10 beta-lactamase, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K54
 
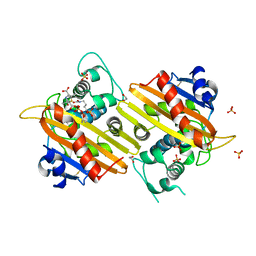 | | OXA-10 class D beta-lactamase partially acylated with reacted 6beta-(1-hydroxy-1-methylethyl) penicillanic acid | | Descriptor: | (1R)-2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Beta lactamase OXA-10, ... | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2QPN
 
 | | GES-1 beta-lactamase | | Descriptor: | Beta-lactamase GES-1, SULFATE ION | | Authors: | Smith, C.A, Caccamo, M, Kantardjieff, K.A, Vakulenko, S. | | Deposit date: | 2007-07-24 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of GES-1 at atomic resolution: insights into the evolution of carbapenamase activity in the class A extended-spectrum beta-lactamases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1E3U
 
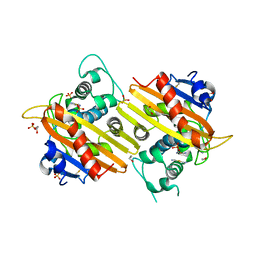 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1E4D
 
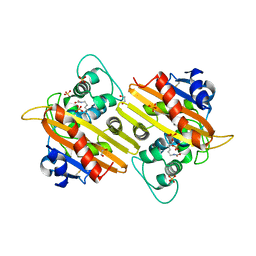 | | Structure of OXA10 beta-lactamase at pH 8.3 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, SULFATE ION | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-07-03 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
4K91
 
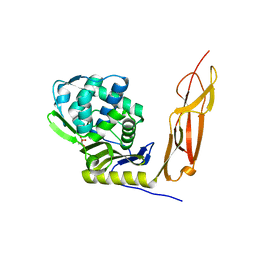 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
