1V8X
 
 | | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae | | Descriptor: | Heme oxygenase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Chu, G.C, Couture, M, Yoshida, T, Rousseau, D.L, Olson, J.S, Ikeda-Saito, M. | | Deposit date: | 2004-01-15 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae: IMPLICATIONS FOR HEME OXYGENASE FUNCTION.
J.Biol.Chem., 279, 2004
|
|
7YL8
 
 | |
7YK9
 
 | | Neutron Structure of PcyA I86D Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Unno, M, Igarashi, K. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YKB
 
 | | Neutron Structure of PcyA D105N Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Unno, M, Nanasawa, R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.38 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
3QXM
 
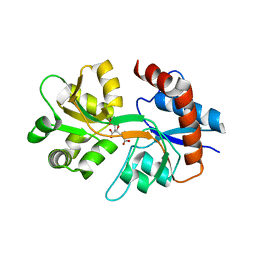 | | Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4QCD
 
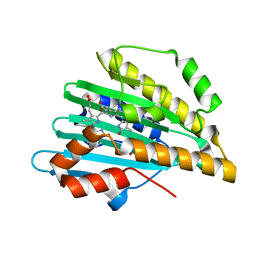 | | Neutron crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha at room temperature. | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, trideuteriooxidanium | | Authors: | Unno, M, Ishikawa-Suto, K, Ishihara, M, Hagiwara, Y, Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.932 Å), X-RAY DIFFRACTION | | Cite: | Insights into the Proton Transfer Mechanism of a Bilin Reductase PcyA Following Neutron Crystallography.
J. Am. Chem. Soc., 137, 2015
|
|
4J20
 
 | | X-ray structure of the cytochrome c-554 from chlorobaculum tepidum | | Descriptor: | Cytochrome c-555, HEME B/C, ISOPROPYL ALCOHOL, ... | | Authors: | Unno, M, Yu, L.J, Wang-otomo, Z.Y. | | Deposit date: | 2013-02-04 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure analysis and characterization of the cytochrome c-554 from thermophilic green sulfur photosynthetic bacterium Chlorobaculum tepidum
Photosynth.Res., 118, 2013
|
|
3NSO
 
 | |
7YRK
 
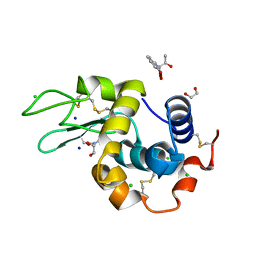 | | Crystal structure of the hen egg lysozyme-2-oxidobenzylidene-threoninato-copper (II) complex | | Descriptor: | (6~{S})-6-[(1~{R})-1-oxidanylethyl]-2,4-dioxa-7$l^{4}-aza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(13),7,9,11-tetraen-5-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Unno, M, Furuya, T, Kitanishi, K, Akitsu, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Sci Rep, 13, 2023
|
|
1IRU
 
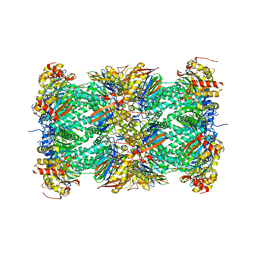 | | Crystal Structure of the mammalian 20S proteasome at 2.75 A resolution | | Descriptor: | 20S proteasome, MAGNESIUM ION | | Authors: | Unno, M, Mizushima, T, Morimoto, Y, Tomisugi, Y, Tanaka, K, Yasuoka, N, Tsukihara, T. | | Deposit date: | 2001-10-24 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of the mammalian 20S proteasome at 2.75 A resolution.
Structure, 10, 2002
|
|
3NSL
 
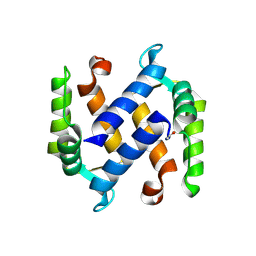 | |
3FVG
 
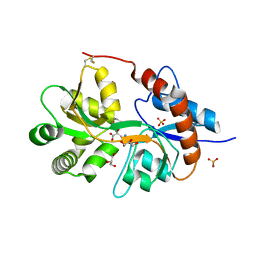 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV1
 
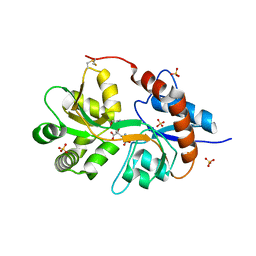 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4GPH
 
 | |
4GPC
 
 | |
4GOH
 
 | |
4GPF
 
 | |
3NSI
 
 | |
3NSK
 
 | |
3FVK
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FUZ
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with L-glutamate in space group P1 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVO
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-epi-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6,7-dihydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Human GluR5 Ligand-Binding Core in Complexes with Novel Marine-Derived Toxins, Dysiherbaine and Neodysiherbaine A, and Their Analogues
To be Published
|
|
3FVN
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 9-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,7R,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-7-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV2
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
1WNX
 
 | | D136E mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO) | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Unno, M, Matsui, T, Ikeda-Saito, M. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM
J.Biol.Chem., 280, 2005
|
|
