1SB3
 
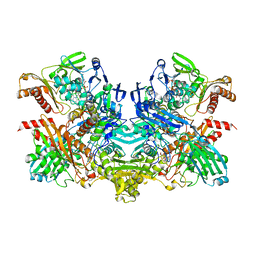 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Boll, M, Ermler, U. | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow.
Structure, 12, 2004
|
|
1RM6
 
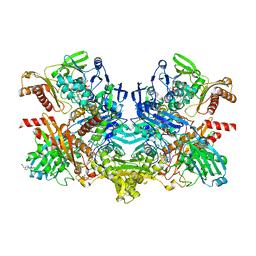 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Dutton, P.L, Boll, M, Ermler, U. | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow
Structure, 12, 2004
|
|
1RGV
 
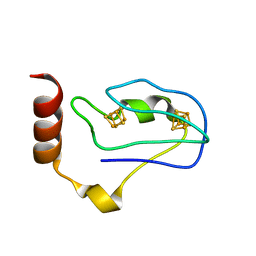 | | Crystal Structure of the Ferredoxin from Thauera aromatica | | Descriptor: | IRON/SULFUR CLUSTER, ferredoxin | | Authors: | Unciuleac, M, Boll, M, Warkentin, E, Ermler, U. | | Deposit date: | 2003-11-13 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5COV
 
 | |
5COT
 
 | |
5COU
 
 | |
6VT4
 
 | | Naegleria gruberi RNA ligase R149A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTE
 
 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT8
 
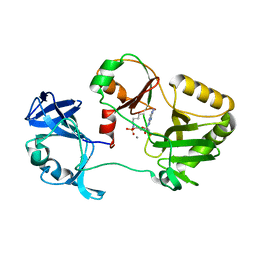 | | Naegleria gruberi RNA ligase E312A mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTD
 
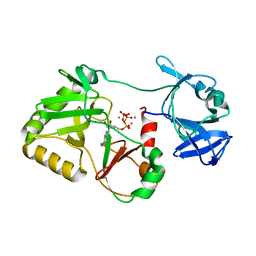 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT6
 
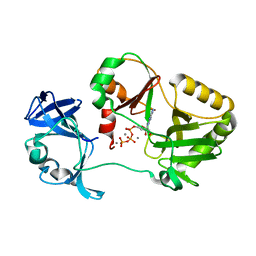 | | Naegleria gruberi RNA ligase K170A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTG
 
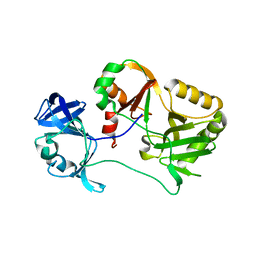 | | Naegleria gruberi RNA ligase E227A mutant apo | | Descriptor: | RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT9
 
 | | Naegleria gruberi RNA ligase E227A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT0
 
 | | Naegleria gruberi RNA ligase K170A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTB
 
 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT1
 
 | | Naegleria gruberi RNA ligase D172A mutant apo | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT5
 
 | |
6VTF
 
 | | Naegleria gruberi RNA ligase with PPi | | Descriptor: | PYROPHOSPHATE 2-, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT3
 
 | | Naegleria gruberi RNA ligase K326A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
7LD5
 
 | | polynucleotide phosphorylase | | Descriptor: | MAGNESIUM ION, Polyribonucleotide nucleotidyltransferase, poly-A RNA fragment | | Authors: | Goldgur, Y, Shuman, S, De La Cruz, M.J, Ghosh, S, Unciuleac, M.-C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and mechanism of Mycobacterium smegmatis polynucleotide phosphorylase.
Rna, 27, 2021
|
|
6NHX
 
 | | mycobacterial DNA ligase D complexed with ATP and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
6NHZ
 
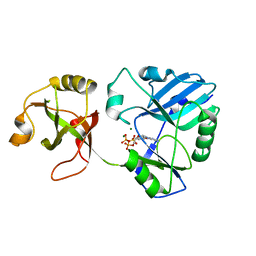 | | mycobacterial DNA ligase D complexed with ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase, MAGNESIUM ION | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
5TT6
 
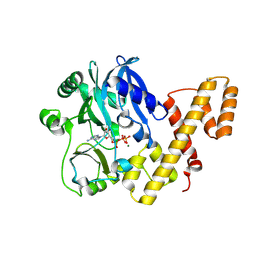 | | T4 RNA Ligase 1 (K99M) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, T4 RNA ligase 1 | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TT5
 
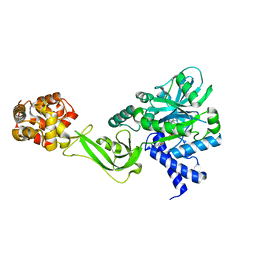 | | Escherichia coli LigA (K115M) in complex with NAD+ | | Descriptor: | DNA ligase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PPR
 
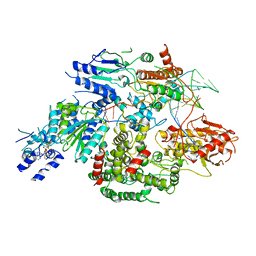 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
