5B0S
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannotriose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0R
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0P
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0Q
 
 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
3WNZ
 
 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alanine-anticapsin ligase BacD, MAGNESIUM ION, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3WO0
 
 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3WO1
 
 | | Crystal structure of Trp332Ala mutant YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3A1E
 
 | | Crystal structure of the P- and N-domains of His462Gln mutant CopA, a copper-transporting P-type ATPase, bound with AMPPCP-Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
3A1C
 
 | | crystal structure of the P- and N-domains of CopA, a copper-transporting P-type ATPase, bound with AMPPCP-Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
3A1D
 
 | | Crystal structure of the P- and N-domains of CopA, a copper-transporting P-type ATPase, bound with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
5YE4
 
 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | Descriptor: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5YE3
 
 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5XWD
 
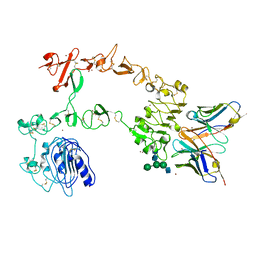 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
1WPG
 
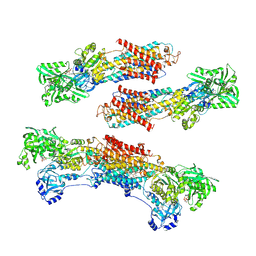 | | Crystal structure of the SR CA2+-ATPase with MGF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T. | | Deposit date: | 2004-09-02 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
2ZBD
 
 | | Crystal Structure of the SR Calcium Pump with Bound Aluminium Fluoride, ADP and Calcium | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2FYH
 
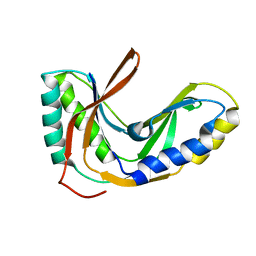 | | Solution structure of the 2'-5' RNA ligase-like protein from Pyrococcus furiosus | | Descriptor: | putative integral membrane transport protein | | Authors: | Okada, K, Matsuda, T, Sakamoto, T, Muto, Y, Yokoyama, S, Kanai, A, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus
Rna, 15, 2009
|
|
1VDY
 
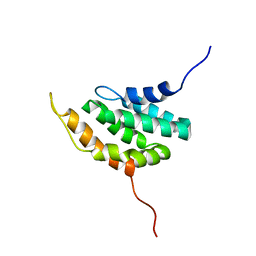 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
1IOZ
 
 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6JXF
 
 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1A
 
 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6ISV
 
 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | Descriptor: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
