7YEC
 
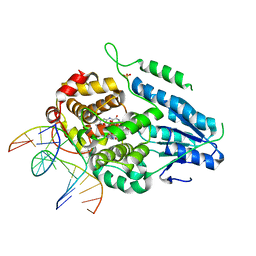 | | TR-SFX MmCPDII-DNA complex: 6 ns snapshot. Includes 6 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEK
 
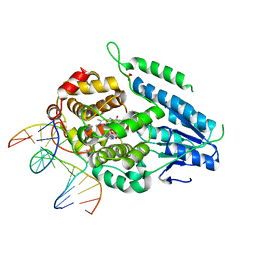 | | TR-SFX MmCPDII-DNA complex: 500 ns time-point collected in SACLA. Includes 500 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEJ
 
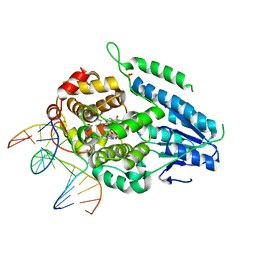 | | TR-SFX MmCPDII-DNA complex: 100 ns time-point collected in SACLA. Includes 100 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEL
 
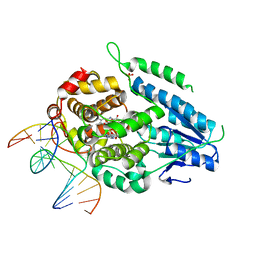 | | TR-SFX MmCPDII-DNA complex: 25 us time-point collected in SACLA. Includes 25 us, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEM
 
 | | TR-SFX MmCPDII-DNA complex: 200 us time-point collected in SACLA. Includes 200 us, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
7CGY
 
 | | Human DMC1 Q244M mutant of the post-synaptic complexes | | Descriptor: | CALCIUM ION, Meiotic recombination protein DMC1/LIM15 homolog, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chi, H.Y, Ho, M.C, Tsai, M.D, Luo, S.C, Yeh, H.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C9C
 
 | | Human DMC1 pre-synaptic complexes | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Meiotic recombination protein DMC1/LIM15 homolog, ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C99
 
 | | Human DMC1 post-synaptic complexes with mismatched dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C9A
 
 | | Human RAD51 post-synaptic complexes mutant (V273P, D274G) | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chi, H.Y, Ho, M.C, Tsai, M.D, Luo, S.C, Yeh, H.Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C98
 
 | | Human DMC1 post-synaptic complexes | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
1VL9
 
 | | Atomic resolution (0.97A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Gayathri, D, Poi, M.-J, Tsai, M.-D, Dauter, M, Dauter, Z. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1GH4
 
 | | Structure of the triple mutant (K56M, K120M, K121M) of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2000-11-09 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Observation of additional calcium ion in the crystal structure of the triple mutant K56,120,121M of bovine pancreatic phospholipase A2.
J.Mol.Biol., 324, 2002
|
|
1O2E
 
 | | Structure of the triple mutant (K53,56,120M) + Anisic acid complex of phospholipase A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the free and anisic acid bound triple mutant of phospholipase A2.
J.Mol.Biol., 333, 2003
|
|
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
8KE2
 
 | | PylRS C-terminus domain mutant bound with L-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2S)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19639969 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE3
 
 | | PylRS C-terminus domain mutant bound with D-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89980984 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE5
 
 | | PylRS C-terminus domain mutant bound with D-3-chlorophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-chlorophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.900073 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE6
 
 | | PylRS C-terminus domain mutant bound with L-3-chlorophenylalanine and AMPNP | | Descriptor: | 3-CHLORO-L-PHENYLALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89570856 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE1
 
 | | PylRS C-terminus domain mutant bound with L-3-bromophenylalanine and AMPNP | | Descriptor: | 3-bromo-L-phenylalanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50081539 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE4
 
 | | PylRS C-terminus domain mutant bound with D-3-bromophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-bromophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75050962 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
7V6H
 
 | | Crystal Structure of the SpnL | | Descriptor: | Cyclopropane fatty-acyl-phospholipid synthase-like methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, H.-H, Ko, T.-P, Liu, H.-W, Tsai, M.-D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Evidence for an Enzyme-Catalyzed Rauhut-Currier Reaction during the Biosynthesis of Spinosyn A.
J.Am.Chem.Soc., 143, 2021
|
|
2BCH
 
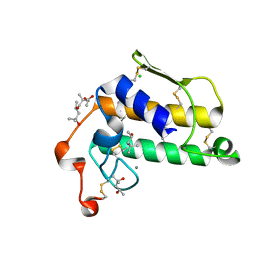 | | A possible of Second calcium ion in interfacial binding: Atomic and Medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Poi, M.J, Dauter, Z, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6A33
 
 | | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA | | Descriptor: | 15-mer peptide from TRAF-interacting protein with FHA domain-containing protein A, TNF receptor-associated factor 6 | | Authors: | Huang, W.C, Liao, J.H, Hsiao, T.C, Maestre-Reyna, M, Bessho, Y, Tsai, M.D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Chembiochem, 20, 2019
|
|
1BVM
 
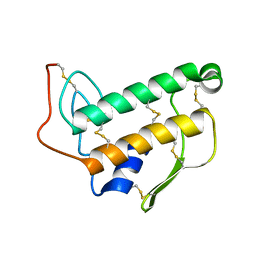 | | SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Yuan, C.-H, Byeon, I.-J.L, Li, Y, Tsai, M.-D. | | Deposit date: | 1998-09-14 | | Release date: | 1999-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of phospholipase A2 from functional perspective. 1. Functionally relevant solution structure and roles of the hydrogen-bonding network.
Biochemistry, 38, 1999
|
|
