1YGH
 
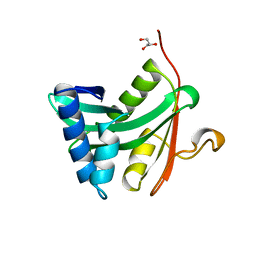 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1P0Y
 
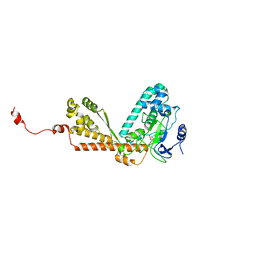 | | Crystal structure of the SET domain of LSMT bound to MeLysine and AdoHcy | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1MLV
 
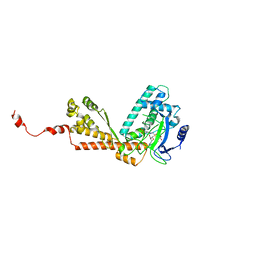 | | Structure and Catalytic Mechanism of a SET Domain Protein Methyltransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ribulose-1,5 biphosphate carboxylase/oxygenase large subunit N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Trievel, R.C, Beach, B.M, Dirk, L.M.A, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a SET domain protein methyltransferase.
Cell(Cambridge,Mass.), 111, 2002
|
|
1OZV
 
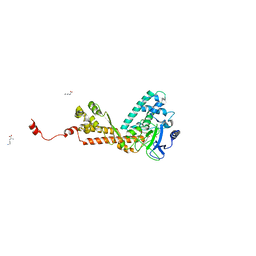 | | Crystal structure of the SET domain of LSMT bound to Lysine and AdoHcy | | Descriptor: | LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-09 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
6M82
 
 | | Crystal structure of TylM1 Y14paF bound to SAH and dTDP-phenol | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.3971 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6M81
 
 | | Crystal structure of TylM1 Y14F bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6M83
 
 | | Crystal structure of TylM1 S120A bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3685 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
5EG2
 
 | | SET7/9 N265A in complex with AdoHcy and TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Kroner, G.M, Fick, R.J, Trievel, R.C. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Acs Chem.Biol., 11, 2016
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
2F69
 
 | | Ternary complex of SET7/9 bound to AdoHcy and a TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Couture, J.-F, Collazo, E, Hauk, G, Trievel, R.C. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the methylation site specificity of SET7/9
Nat.Struct.Mol.Biol., 13, 2006
|
|
1QST
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSN
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
2Q8C
 
 | | Crystal structure of JMJD2A in ternary complex with an histone H3K9me3 peptide and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, HISTONE 3 PEPTIDE, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q8D
 
 | | Crystal structure of JMJ2D2A in ternary complex with histone H3-K36me2 and succinate | | Descriptor: | HISTONE 3 peptide, JmjC domain-containing histone demethylation protein 3A, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q8E
 
 | | Specificity and Mechanism of JMJD2A, a Trimethyllysine-Specific Histone Demethylase | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1ZKK
 
 | | Crystal structure of hSET8 in ternary complex with H4 peptide (16-24) and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase, H4 lysine-20 specific, Peptide corresponding to residues 15-24 of histone H4, ... | | Authors: | Couture, J.-F, Collazo, E, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Genes Dev., 19, 2005
|
|
6BDY
 
 | |
6BM5
 
 | |
6BM6
 
 | |
4J8O
 
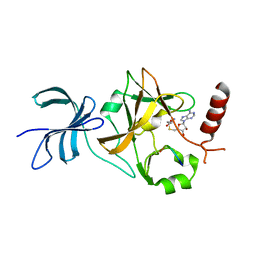 | | SET7/9 in complex with TAF10K189A peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
1CM0
 
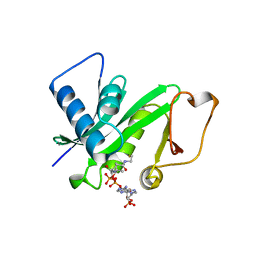 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
4J7F
 
 | |
4J7I
 
 | | SET7/9Y335F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J83
 
 | | SET7/9 in complex with TAF10K189A peptide and AdoMet | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Nimtz, J.S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4JE5
 
 | | Crystal structure of the aromatic aminotransferase Aro8, a putative alpha-aminoadipate aminotransferase in Saccharomyces cerevisiae | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic/aminoadipate aminotransferase 1, ... | | Authors: | Bulfer, S.L, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2013-02-26 | | Release date: | 2013-09-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase.
Protein Sci., 22, 2013
|
|
