8QAJ
 
 | |
7OJ9
 
 | |
1SZL
 
 | | F-spondin TSR domain 1 | | Descriptor: | F-spondin | | Authors: | Tossavainen, H, Paakkonen, K, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
5I22
 
 | | Amphiphysin SH3 in complex with Chikungunya virus nsP3 peptide | | Descriptor: | CHIKV nsP3 peptide, Myc box-dependent-interacting protein 1 | | Authors: | Tossavainen, H, Aitio, O, Hellman, M, Saksela, K, Permi, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-06-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2.
J.Biol.Chem., 291, 2016
|
|
1NYA
 
 | | NMR SOLUTION STRUCTURE OF CALERYTHRIN, AN EF-HAND CALCIUM-BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calerythrin | | Authors: | Tossavainen, H, Permi, P, Annila, A, Kilpelainen, I, Drakenberg, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of calerythrin, an EF-hand calcium-binding protein from Saccharopolyspora erythraea
Eur.J.Biochem., 270, 2003
|
|
5KQC
 
 | |
5NMY
 
 | |
5N9Q
 
 | | Structure of A. thaliana RCD1(468-567) | | Descriptor: | Inactive poly [ADP-ribose] polymerase RCD1 | | Authors: | Tossavainen, H, Hellman, M, Vainonen, J, Kangasjarvi, J, Permi, P. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors.
Elife, 8, 2019
|
|
2BBX
 
 | |
1ZK6
 
 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
2MF6
 
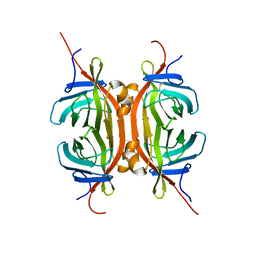 | | Solution NMR structure of Chimeric Avidin, ChiAVD(I117Y), in the biotin bound form | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Tossavainen, H, Kukkurainen, S, Maatta, J.A.E, Pihlajamaa, T, Hytonen, V.P, Kulomaa, M.S, Permi, P. | | Deposit date: | 2013-10-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chimeric Avidin - NMR Structure and Dynamics of a 56 kDa Homotetrameric Thermostable Protein
Plos One, 9, 2014
|
|
2N1U
 
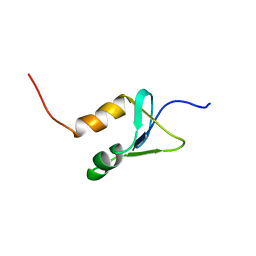 | | Structure of SAP30L corepressor protein | | Descriptor: | Histone deacetylase complex subunit SAP30L, ZINC ION | | Authors: | Tossavainen, H, Permi, P. | | Deposit date: | 2015-04-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function.
Protein Sci., 25, 2016
|
|
1VEX
 
 | | F-spondin TSR domain 4 | | Descriptor: | F-spondin | | Authors: | Paakkonen, K, Tossavainen, H, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
5KQB
 
 | |
2V6Z
 
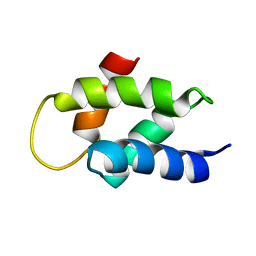 | | Solution Structure of Amino-Terminal Domain of Human DNA Polymerase Epsilon Subunit B | | Descriptor: | DNA POLYMERASE EPSILON SUBUNIT 2 | | Authors: | Nuutinen, T, Fredriksson, K, Tossavainen, H, Pospiech, H, Pirila, P, Permi, P, Annila, A, Syvaoja, J.E. | | Deposit date: | 2007-07-24 | | Release date: | 2008-08-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Amino-Terminal Domain of Human DNA Polymerase Epsilon Subunit B is Homologous to C-Domains of Aaa+ Proteins.
Nucleic Acids Res., 36, 2008
|
|
5N9V
 
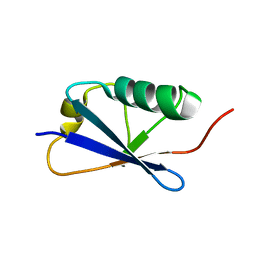 | |
2UUR
 
 | | N-terminal NC4 domain of collagen IX | | Descriptor: | 1,2-ETHANEDIOL, COLLAGEN ALPHA-1(IX) CHAIN, SULFATE ION, ... | | Authors: | Leppanen, V.-M, Tossavainen, H, Permi, P, Lehtio, L, Ronnholm, G, Goldman, A, Kilpelainen, I, Pihlajamaa, T. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Terminal Nc4 Domain of Collagen Ix, a Zinc Binding Member of the Laminin-Neurexin-Sex Hormone Binding Globulin (Lns) Domain Family.
J.Biol.Chem., 282, 2007
|
|
2ROL
 
 | | Structural Basis of PxxDY motif recognition in SH3 binding | | Descriptor: | 12-meric peptide from T-cell surface glycoprotein CD3 epsilon chain, Epidermal growth factor receptor kinase substrate 8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Paakkonen, K, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding
J.Mol.Biol., 382, 2008
|
|
2ALB
 
 | |
2JZV
 
 | | Solution structure of S. aureus PrsA-PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Seppala, R, Tossavainen, H, Heikkinen, S, Koskela, H, Kontinen, V, Permi, P. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA - Implications for the catalytic mechanism of parvulins.
Bmc Struct.Biol., 9, 2009
|
|
2K2M
 
 | | Structural Basis of PxxDY Motif Recognition in SH3 Binding | | Descriptor: | Eps8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding.
J.Mol.Biol., 382, 2008
|
|
