3VC6
 
 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with magnesium and formate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
3VC5
 
 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with phosphate | | Descriptor: | Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
3VDG
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
3VFC
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with tartrate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 from Mycobacterium smegmatis
To be Published
|
|
3V7P
 
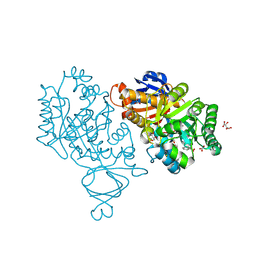 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
4YHS
 
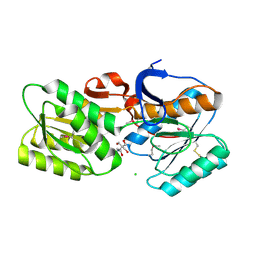 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS
To be published
|
|
4YIC
 
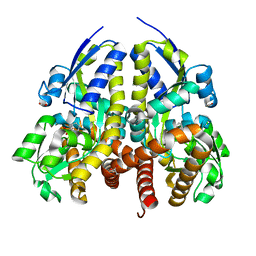 | | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID | | Descriptor: | ACETATE ION, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-01 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID
To be published
|
|
4YQY
 
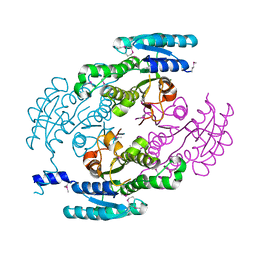 | | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028) (TARGET EFI-513936) in its APO form | | Descriptor: | MAGNESIUM ION, Putative Dehydrogenase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028, TARGET EFI-513936) in its APO form
To be published
|
|
4YWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE | | Descriptor: | ABC TRANSPORTER SOLUTE BINDING PROTEIN, beta-D-xylopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE
To be published
|
|
4YS6
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
3TJI
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ENTEROBACTER sp. 638 (EFI TARGET EFI-501662) with BOUND MG | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from enterobacter sp. 638 (efi target efi-501662) with boung mg
to be published
|
|
4YLE
 
 | | Crystal structure of an ABC transpoter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-15 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose
To be published
|
|
4YQZ
 
 | | Crystal Structure of a putative oxidoreductase from Thermus Thermophilus HB27 (TT_P0034, TARGET EFI-513932) in its APO form | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Crystal Structure of a putative oxidoreductase from Thermus Thermophilus HB27 (TT_P0034, TARGET EFI-513932) in its APO form
To be published
|
|
4YO7
 
 | | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-11 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol
To be published
|
|
4YV7
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL | | Descriptor: | GLYCEROL, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL
To be published
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
4ZJP
 
 | | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose | | Descriptor: | 1,2-ETHANEDIOL, Monosaccharide-transporting ATPase, beta-D-ribopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Glenn, A.S, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
To be published
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | Descriptor: | Putative periplasmic binding protein with substrate ribose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
5CM6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE
To be published
|
|
3UIF
 
 | | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein SsuA from Methylobacillus flagellatus KT | | Descriptor: | GLYCEROL, SULFATE ION, Sulfonate ABC transporter, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein
SsuA from Methylobacillus flagellatus KT
To be Published
|
|
3UOG
 
 | | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Alcohol dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3V3W
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol
to be published
|
|
3V4B
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
