3CTP
 
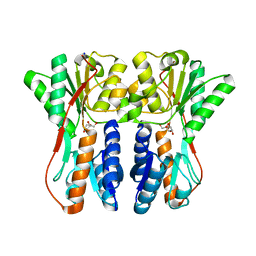 | | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with D-xylulofuranose | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, SODIUM ION, beta-D-xylulofuranose | | 著者 | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-14 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
To be Published
|
|
3P9X
 
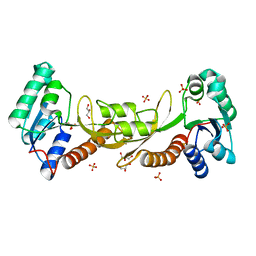 | | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans | | 分子名称: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyltransferase | | 著者 | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2010-10-18 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans
To be Published
|
|
3DFH
 
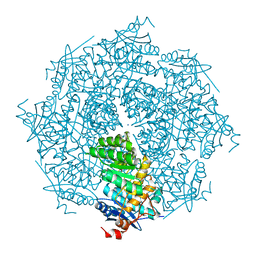 | | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3 | | 分子名称: | SODIUM ION, mandelate racemase | | 著者 | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-06-12 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3
To be Published
|
|
2YB4
 
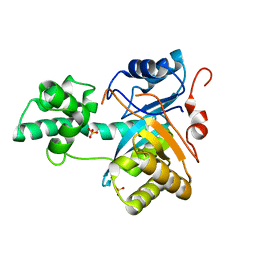 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | 分子名称: | AMIDOHYDROLASE, SULFATE ION | | 著者 | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-03-01 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
2YB1
 
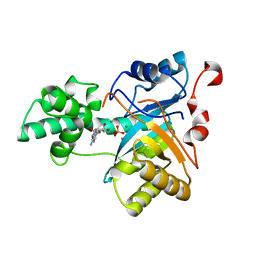 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound Mn, AMP and phosphate. | | 分子名称: | ADENOSINE MONOPHOSPHATE, AMIDOHYDROLASE, MANGANESE (II) ION, ... | | 著者 | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-02-25 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
3EBV
 
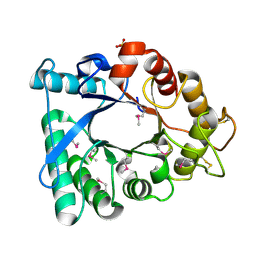 | | Crystal structure of putative Chitinase A from Streptomyces coelicolor. | | 分子名称: | Chinitase A, SULFATE ION | | 著者 | Vigdorovich, V, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-08-28 | | 公開日 | 2008-09-30 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of putative Chitinase A from Streptomyces coelicolor
To be Published
|
|
4DLM
 
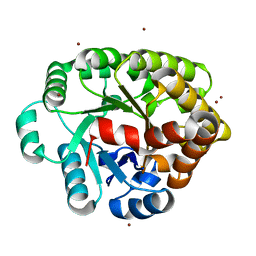 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | 分子名称: | Amidohydrolase 2, ZINC ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
4DX3
 
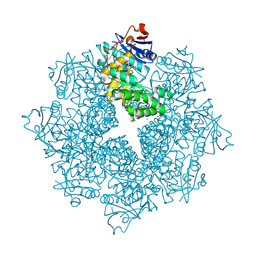 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue | | 分子名称: | CHLORIDE ION, GLYCEROL, Mandelate racemase / muconate lactonizing enzyme family protein | | 著者 | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-27 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
to be published
|
|
4DLF
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P3221 | | 分子名称: | Amidohydrolase 2, GLYCEROL, UNKNOWN ATOM OR ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (target efi-500235) with bound ZN, space group P3221
to be published
|
|
4DN1
 
 | | Crystal structure of an ENOLASE (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate | | 分子名称: | CHLORIDE ION, FORMIC ACID, Isomerase/lactonizing enzyme, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Sojitra, S, Al Obaidi, N.F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-08 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of an enolase (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate
to be published
|
|
3EGO
 
 | | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis | | 分子名称: | Probable 2-dehydropantoate 2-reductase | | 著者 | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-09-11 | | 公開日 | 2008-09-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis
To be published
|
|
4DNM
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase 2, GLYCEROL, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-08 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221
to be published
|
|
3QAN
 
 | | Crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from bacillus halodurans | | 分子名称: | 1-pyrroline-5-carboxylate dehydrogenase 1, ACETATE ION | | 著者 | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-01-11 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of 1-Pyrroline-5-Carboxylate Dehydrogenase from Bacillus Halodurans
To be Published
|
|
3EPR
 
 | | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae. | | 分子名称: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family, ... | | 著者 | Ramagopal, U.A, Toro, R, Dickey, M, Tang, B.K, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-09-29 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae.
To be Published
|
|
4DXK
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate | | 分子名称: | Mandelate racemase / muconate lactonizing enzyme family protein, PHOSPHATE ION, SODIUM ION | | 著者 | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-27 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate
to be published
|
|
3EVN
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Streptococcus agalactiae 2603V/r | | 分子名称: | Oxidoreductase, Gfo/Idh/MocA family | | 著者 | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-10-13 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF putative Gfo/Idh/MocA family oxidoreductase from Streptococcus agalactiae
2603V/r
To be Published
|
|
3QU9
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with magnesium and tartrate | | 分子名称: | CHLORIDE ION, GLYCEROL, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
4ESO
 
 | | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | 著者 | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-04-23 | | 公開日 | 2012-05-16 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP
To be Published
|
|
3FEF
 
 | | Crystal structure of putative glucosidase lplD from bacillus subtilis | | 分子名称: | MAGNESIUM ION, Putative glucosidase lplD, ALPHA-GALACTURONIDASE, ... | | 著者 | Ramagopal, U.A, Rajashankar, K.R, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-11-28 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of putative glucosidase lplD from bacillus subtilis.
To be published
|
|
3QXG
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron complexed with calcium, a closed cap conformation | | 分子名称: | ACETATE ION, CALCIUM ION, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-03-01 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU4
 
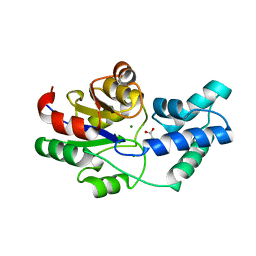 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13ala mutant | | 分子名称: | ACETATE ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUB
 
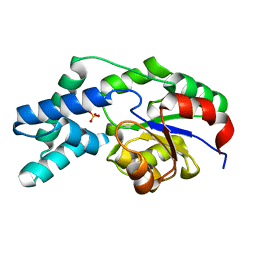 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47ala mutant complexed with sulfate | | 分子名称: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QX7
 
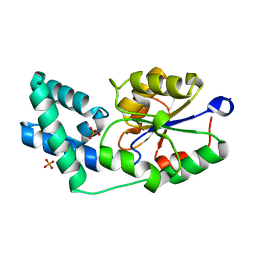 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron complexed with phosphate, a closed cap conformation | | 分子名称: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-03-01 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUQ
 
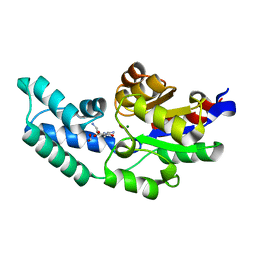 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, an open cap conformation | | 分子名称: | CHLORIDE ION, FORMIC ACID, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-24 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU5
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp11asn mutant | | 分子名称: | CHLORIDE ION, INORGANIC PYROPHOSPHATASE | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
