6Z7O
 
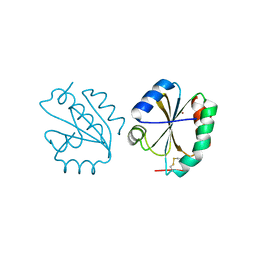 | | Crystal structure of Thioredoxin T from Drosophila melanogaster | | Descriptor: | Thioredoxin-T, ZINC ION | | Authors: | Freier, R, Aragon, E, Baginski, B, Pluta, R, Martin-Malpartida, P, Torner, C, Gonzaez, C, Macias, M. | | Deposit date: | 2020-05-31 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
8RTH
 
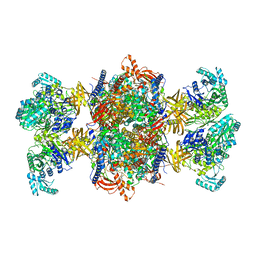 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 32, 2024
|
|
6TUX
 
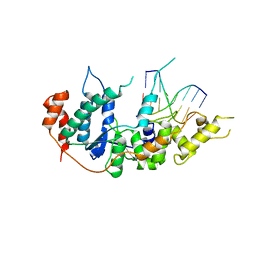 | | human XPG-DNA, Complex 2 | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*AP*GP*TP*T)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
6TUR
 
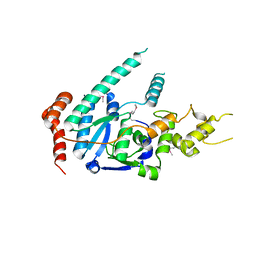 | | human XPG, Apo1 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
8BWS
 
 | | Structure of yeast RNA Polymerase III elongation complex at 3.3 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
6TUW
 
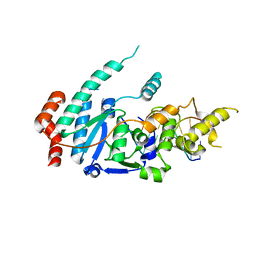 | | human XPG-DNA, Complex 1 | | Descriptor: | DNA (5'-D(P*GP*AP*AP*CP*TP*CP*TP*G)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*AP*GP*TP*TP*C)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
3NSU
 
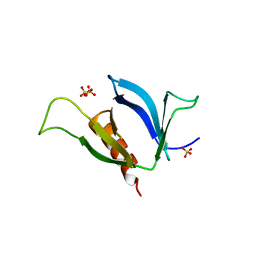 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
5LMX
 
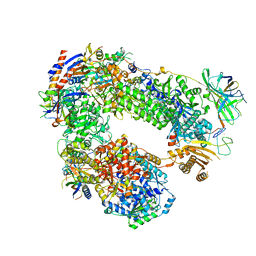 | | Monomeric RNA polymerase I at 4.9 A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Torreira, E, Louro, J.A, Gil-Carton, D, Gallego, O, Calvo, O, Fernandez-Tornero, C. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription.
Elife, 6, 2017
|
|
2J04
 
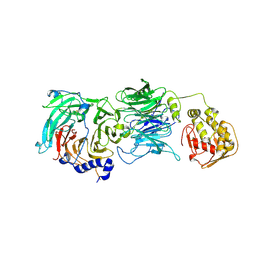 | | The tau60-tau91 subcomplex of yeast transcription factor IIIC | | Descriptor: | HYPOTHETICAL PROTEIN YPL007C, YDR362CP | | Authors: | Mylona, A, Fernandez-Tornero, C, Legrand, P, Muller, C.W. | | Deposit date: | 2006-07-31 | | Release date: | 2006-10-23 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Tau60/Deltatau91 Subcomplex of Yeast Transcription Factor Iiic: Insights Into Preinitiation Complex Assembly
Mol.Cell, 24, 2006
|
|
6TUS
 
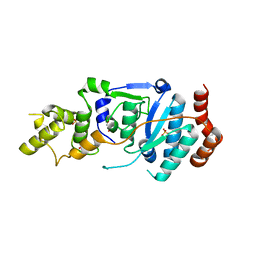 | | human XPG, Apo2 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells, GLYCEROL, SULFATE ION | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
6YD1
 
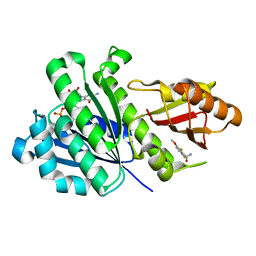 | | SaFtsZ-DFMBA | | Descriptor: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD6
 
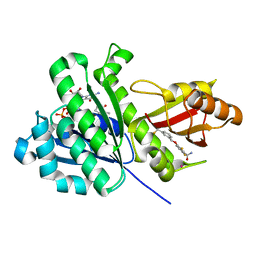 | | SaFtsZ-UCM152 (comp.20) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[3-(trifluoromethyl)phenyl]methoxy]benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD5
 
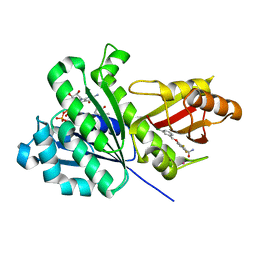 | | SaFtsZ-UCM151 (comp. 18) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
7Z31
 
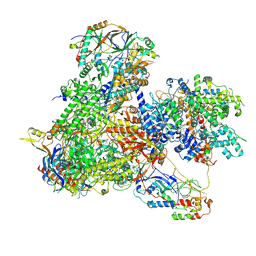 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
2YN0
 
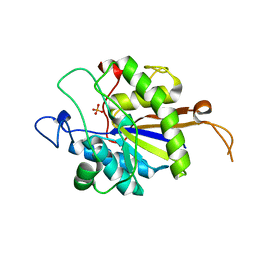 | | tau55 histidine phosphatase domain | | Descriptor: | PHOSPHATE ION, TRANSCRIPTION FACTOR TAU 55 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Iiic.
J.Biol.Chem., 288, 2013
|
|
7ON3
 
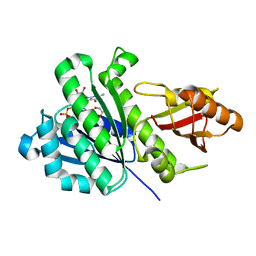 | | SaFtsZ complexed with GDP (soak 10 mM EGTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMP
 
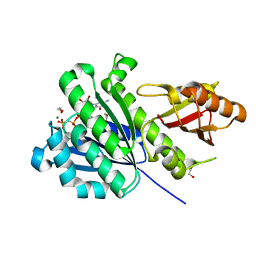 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7ON4
 
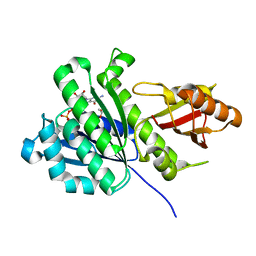 | | SaFtsZ complexed with GDP (co-crystalization with 1mM EDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMQ
 
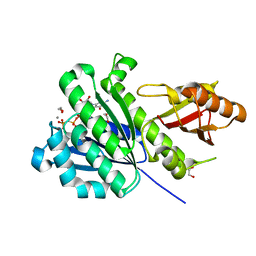 | | SaFtsZ complexed with GDPPCP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MANGANESE (II) ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7ON2
 
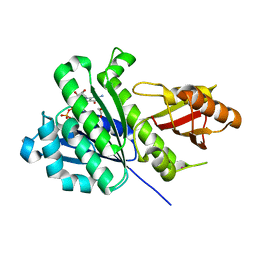 | | SaFtsZ complexed with GDP (soaking in 10 mM CyDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMJ
 
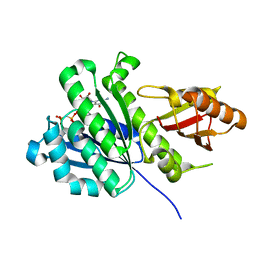 | | SaFtsZ complexed with GDPPCP | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
2BYM
 
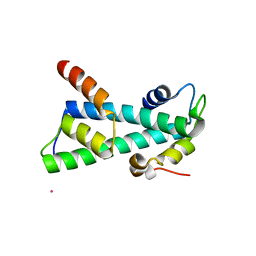 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CADMIUM ION, CHRAC-14, CHRAC-16 | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
2BYK
 
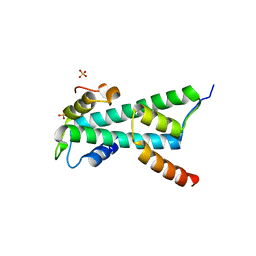 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CHRAC-14, CHRAC-16, SULFATE ION | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
6RVM
 
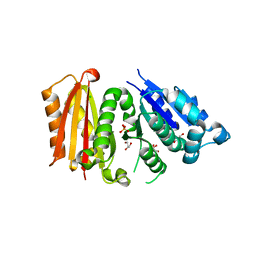 | | Cell division protein FtsZ from Staphylococcus aureus, apo form | | Descriptor: | CHLORIDE ION, Cell division protein FtsZ, GLYCEROL, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Canosa-Valls, A.J. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
7Z0H
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
