7ZOA
 
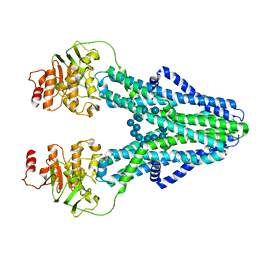 | | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, Cyclooctadecakis-(1-2)-(beta-D-glucopyranose) | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1NX2
 
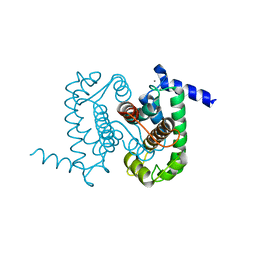 | | Calpain Domain VI | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
5V8Y
 
 | |
5JYF
 
 | |
2V54
 
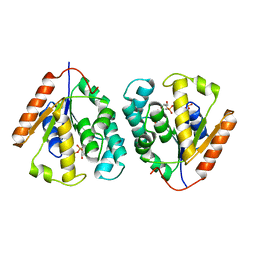 | | Crystal structure of vaccinia virus thymidylate kinase bound to TDP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1DOJ
 
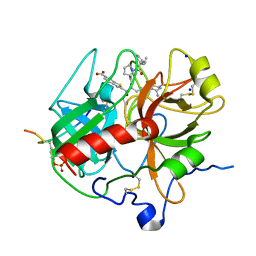 | | Crystal structure of human alpha-thrombin*RWJ-51438 complex at 1.7 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUGEN, ... | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Carson, M, DeLucas, L, Chattopadhyay, D. | | Deposit date: | 1999-12-21 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human alpha-thrombin complexed with RWJ-51438 at 1.7 A: unusual perturbation of the 60A-60I insertion loop.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1NX3
 
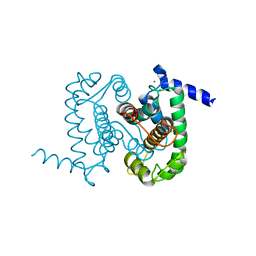 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1ALV
 
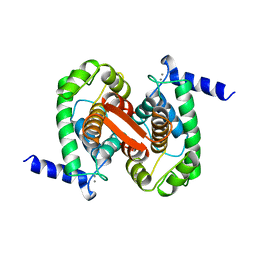 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1VSU
 
 | |
1QCP
 
 | | CRYSTAL STRUCTURE OF THE RWJ-51084 BOVINE PANCREATIC BETA-TRYPSIN AT 1.8 A | | Descriptor: | CALCIUM ION, CYCLOPENTANECARBOXYLIC ACID [1-(BENZOTHIAZOLE-2-CARBONYL)-4-GUANIDINO-BUTYL]-AMIDE, PROTEIN (BETA-TRYPSIN PROTEIN) | | Authors: | Recacha, R, Carson, M, Costanzo, M.J, Maryanoff, B, Chattopadhyay, D. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RWJ-51084-bovine pancreatic beta-trypsin complex at 1.8 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6X2E
 
 | |
5UTM
 
 | |
4QCA
 
 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant R167AD4 | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of three recombinant mutants of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6WYC
 
 | |
5UTL
 
 | |
5V8X
 
 | |
1MXH
 
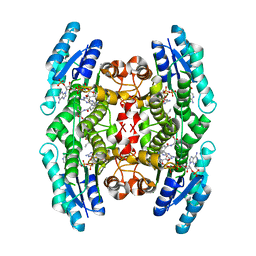 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
4DRS
 
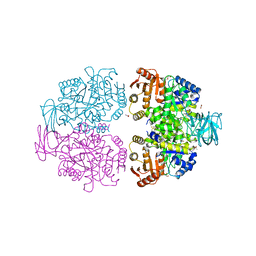 | |
4DOG
 
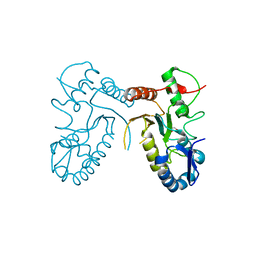 | |
1MXF
 
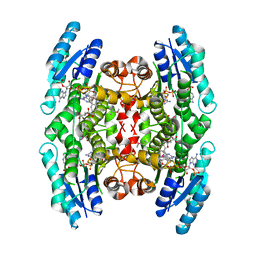 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
4QX6
 
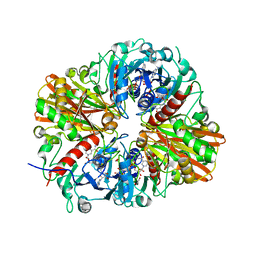 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5MGM
 
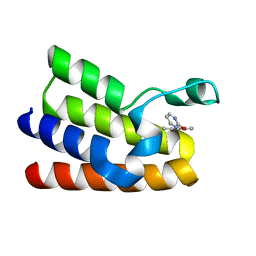 | |
4QCB
 
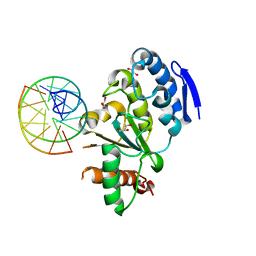 | | Protein-DNA complex of Vaccinia virus D4 with double-stranded non-specific DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*C)-3', GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Banerjee, S, Ricciardi, R, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Binding of undamaged double stranded DNA to vaccinia virus uracil-DNA Glycosylase.
BMC Struct. Biol., 15, 2015
|
|
4QC9
 
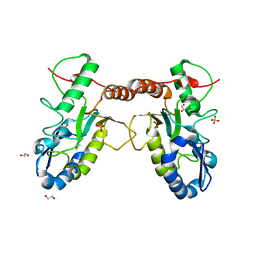 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
4LZB
 
 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
