6DNH
 
 | | Cryo-EM structure of human CPSF-160-WDR33-CPSF-30-PAS RNA complex at 3.4 A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*C)-3'), ... | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1QYI
 
 | | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25. | | Descriptor: | hypothetical protein | | Authors: | Kuzin, A.P, Edstrom, W, Ma, L.C, Shin, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-10 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25.
To be Published
|
|
1EFL
 
 | | HUMAN MALIC ENZYME IN A QUATERNARY COMPLEX WITH NAD, MG, AND TARTRONATE | | Descriptor: | MAGNESIUM ION, MALIC ENZYME, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
3BG9
 
 | |
1EFK
 
 | | STRUCTURE OF HUMAN MALIC ENZYME IN COMPLEX WITH KETOMALONATE | | Descriptor: | ALPHA-KETOMALONIC ACID, MAGNESIUM ION, MALIC ENZYME, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
4HNV
 
 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNT
 
 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
6AOZ
 
 | |
2OKA
 
 | | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR82 | | Descriptor: | Hypothetical protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Chen, X.C, Fang, Y, Cunningham, K, Owens, L, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-16 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa
To be Published
|
|
5BTH
 
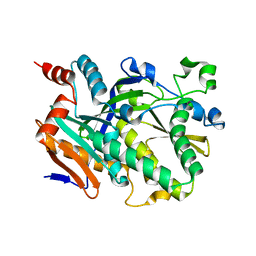 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BTO
 
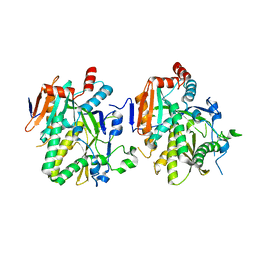 | |
5BTB
 
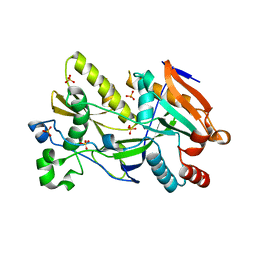 | | Crystal Structure of Ashbya gossypii Rai1 | | Descriptor: | AFR263Cp, SULFATE ION | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
1IEC
 
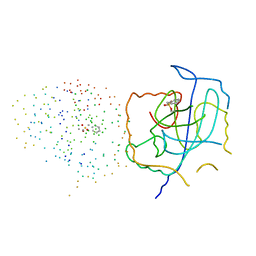 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157A) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A/H157A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEF
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IED
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157E) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1PJ2
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
5CSL
 
 | |
5CS4
 
 | |
5VIT
 
 | |
1KV2
 
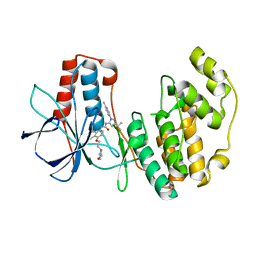 | | Human p38 MAP Kinase in Complex with BIRB 796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
1KV1
 
 | | p38 MAP Kinase in Complex with Inhibitor 1 | | Descriptor: | 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
5CSA
 
 | |
4HNU
 
 | | crystal structure of K442E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
5CS0
 
 | |
