8S9S
 
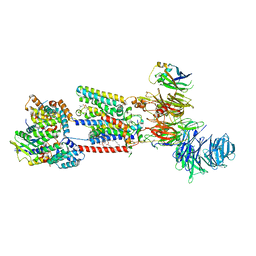 | | Structure of the human ER membrane protein complex (EMC) in GDN | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tomaleri, G.P, Nguyen, V.N, Voorhees, R.M. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER.
J.Cell Biol., 222, 2023
|
|
6WW7
 
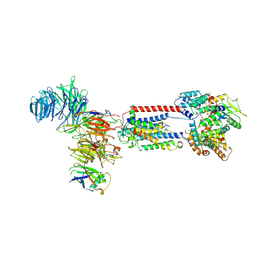 | | Structure of the human ER membrane protein complex in a lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ER Membrane Protein Complex Subunit 4, ... | | Authors: | Tomaleri, G.P, Januszyk, K, Pleiner, T, Inglis, A.J, Voorhees, R.M. | | Deposit date: | 2020-05-08 | | Release date: | 2020-05-27 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for membrane insertion by the human ER membrane protein complex.
Science, 369, 2020
|
|
9C7U
 
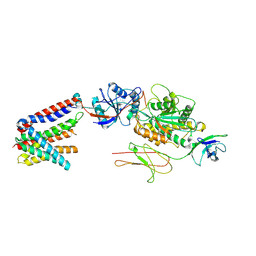 | | Structure of the human truncated BOS complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, Nicalin, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
9C7V
 
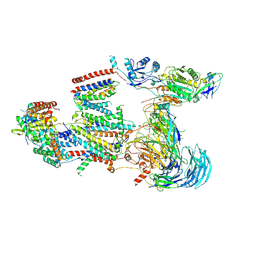 | | Structure of the human BOS:human EMC complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
7TBK
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) symmetric core generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | Descriptor: | NUP107 CTD, NUP107 NTD, NUP133, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBI
 
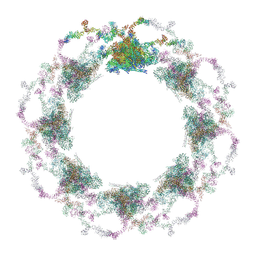 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | Descriptor: | Dyn2, Nic96 R1, Nic96 R2, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | NUP107 CTD, NUP107 NTD, NUP133, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5VL9
 
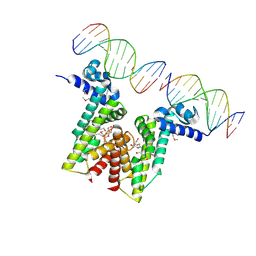 | | Crystal structure of EilR in complex with eilO DNA element | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*GP*TP*TP*GP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*CP*AP*AP*CP*TP*TP*TP*C)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
5VLG
 
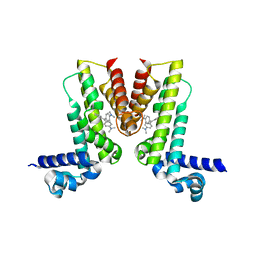 | | Crystal structure of EilR in complex with malachite green | | Descriptor: | MALACHITE GREEN, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.932 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
5VLM
 
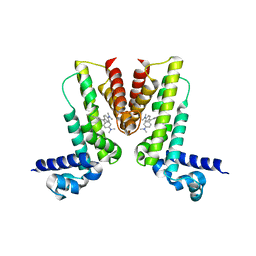 | | Crystal structure of EilR in complex with crystal violet | | Descriptor: | CRYSTAL VIOLET, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
7MVW
 
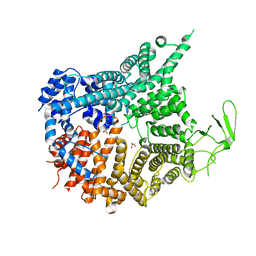 | | Crystal structure of Chaetomium thermophilum Nup188 NTD (residues 1-1134) | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVT
 
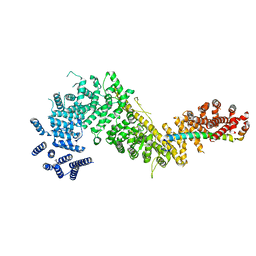 | | Crystal structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 185-1756; Nic96 residues 187-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVV
 
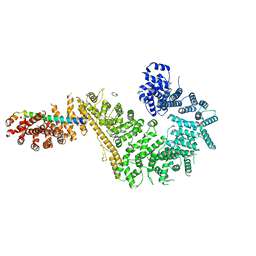 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96-Nup53-Nup145N complex (Nup192 residues 1-1756; Nic96 residues 240-301; Nup53 31-67; Nup145N 616-683) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP192, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVZ
 
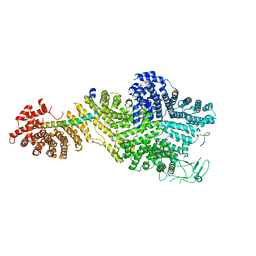 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96-Nup145N complex (Nup188 residues 1-1858; Nic96 residues 240-301; Nup145N residues 640-732) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVX
 
 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW0
 
 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW1
 
 | | Crystal structure of the Homo sapiens NUP93-NUP53 complex (NUP93 residues 174-819; NUP53 residues 84-150) | | Descriptor: | Nuclear pore complex protein Nup93, Nucleoporin Nup35 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVU
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 1-1756; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVY
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
