8BA1
 
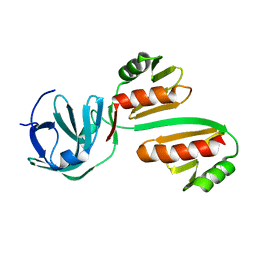 | | CTD12-CTD12 heterodimer from CPSF73 and CPSF100 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
8B7T
 
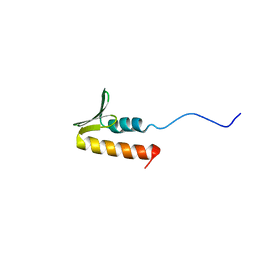 | | CPSF73 CTD3 | | Descriptor: | CPSF73 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
2JJ6
 
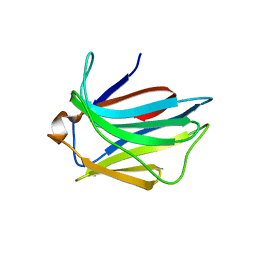 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
5NLG
 
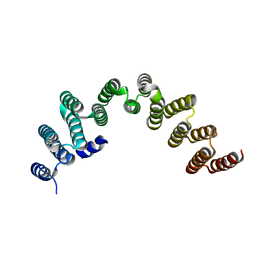 | | RRP5 C-terminal domain | | Descriptor: | rRNA biogenesis protein RRP5 | | Authors: | Thore, S, Fribourg, S. | | Deposit date: | 2017-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and interaction analysis of the Rrp5 C-terminal region.
FEBS Open Bio, 8, 2018
|
|
1M8V
 
 | | Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', CALCIUM ION, PUTATIVE SNRNP SM-LIKE PROTEIN, ... | | Authors: | Thore, S, Mayer, C, Sauter, C, Weeks, S, Suck, D. | | Deposit date: | 2002-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pyrococcus abyssii Sm core and its Complex with RNA: Common Features of RNA-binding in Archaea and Eukarya
J.Biol.Chem., 278, 2003
|
|
2CKY
 
 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
1UOC
 
 | | X-ray structure of the RNase domain of the yeast Pop2 protein | | Descriptor: | CALCIUM ION, POP2, XENON | | Authors: | Thore, S, Mauxion, F, Seraphin, B, Suck, D. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure and Activity of the Yeast Pop2 Protein: A Nuclease Subunit of the Mrna Deadenylase Complex
Embo Rep., 4, 2003
|
|
3D2X
 
 | | Structure of the thiamine pyrophosphate-specific riboswitch bound to oxythiamine pyrophosphate | | Descriptor: | 3-[(4-hydroxy-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, MAGNESIUM ION, TPP-specific riboswitch | | Authors: | Thore, S, Frick, C, Ban, N. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch
J.Am.Chem.Soc., 130, 2008
|
|
3D2G
 
 | |
3D2V
 
 | |
5HZD
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei | | Descriptor: | 3' terminal uridylyl transferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5I49
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP analog UMPNPP | | Descriptor: | 3' terminal uridylyl transferase, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5IDO
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
1I2Q
 
 | |
1I2O
 
 | |
1I2N
 
 | |
1I2P
 
 | |
1I2R
 
 | |
6GIQ
 
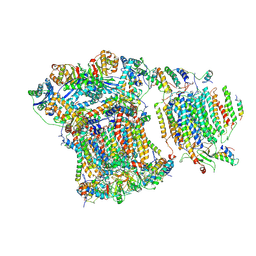 | | Saccharomyces cerevisiae respiratory supercomplex III2IV | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Rathore, S, Berndtsson, J, Conrad, J, Ott, M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the yeast respiratory supercomplex.
Nat. Struct. Mol. Biol., 26, 2019
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
2R0H
 
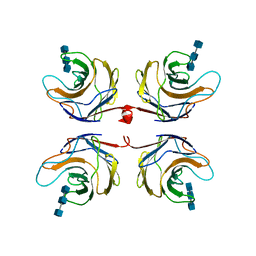 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-20 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
2R0F
 
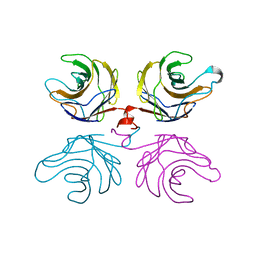 | | Ligand free structure of fungal lectin CGL3 | | Descriptor: | CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1I4K
 
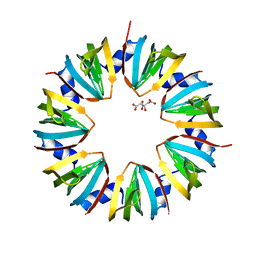 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
4WN4
 
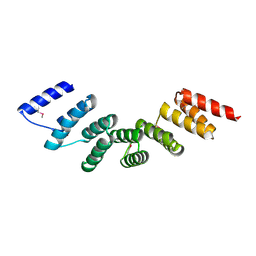 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
