1LRL
 
 | | Crystal Structure of UDP-Galactose 4-Epimerase Mutant Y299C Complexed with UDP-Glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
1LO9
 
 | | X-ray crystal structure of 4-hydroxybenzoyl CoA thioesterase mutant D17N complexed with 4-hydroxybenzoyl CoA | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1LRJ
 
 | | Crystal Structure of E. coli UDP-Galactose 4-Epimerase Complexed with UDP-N-Acetylglucosamine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
1LRK
 
 | | Crystal Structure of Escherichia coli UDP-Galactose 4-Epimerase Mutant Y299C Complexed with UDP-N-acetylglucosamine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
1MUH
 
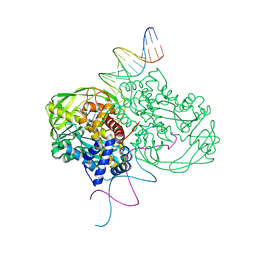 | | CRYSTAL STRUCTURE OF TN5 TRANSPOSASE COMPLEXED WITH TRANSPOSON END DNA | | Descriptor: | DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Davies, D.R, Goryshin, I.Y, Reznikoff, W.S, Rayment, I. | | Deposit date: | 2002-09-23 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the Tn5 synaptic complex transposition intermediate.
Science, 289, 2000
|
|
1JXZ
 
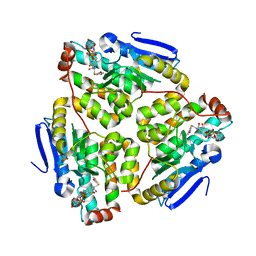 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
5T6B
 
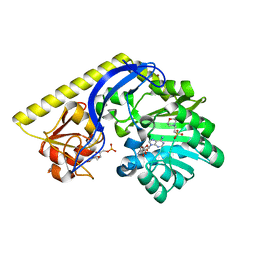 | | X-ray structure of the KijD1 C3-methyltransfeerase, converted to monomeric form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, Sugar 3-C-methyl transferase, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T. | | Deposit date: | 2016-09-01 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies on KijD1, a sugar C-3'-methyltransferase.
Protein Sci., 25, 2016
|
|
8E77
 
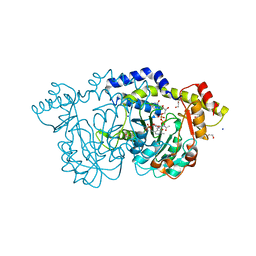 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E75
 
 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E62
 
 | | STRUCTURE OF Pcryo_0615 from Psychrobacter cryohalolentis, an N-acetyltransferase required to produce Diacetamido-2,3-dideoxy-D-glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, COENZYME A, SODIUM ION, ... | | Authors: | Hofmeister, D.L, Bockhaus, N.J, Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
7M15
 
 | | crystal structure of cj1430 in the presence of GDP-D-glycero-L-gluco-heptose, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-L-gluco-heptose, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4R,5R,6S)-6-[(1R)-1,2-dihydroxyethyl]-3,4,5-trihydroxyoxan-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M13
 
 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M14
 
 | | x-ray structure of cj1430 in the presence of GDP, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7MFO
 
 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFP
 
 | | crystal structure of the L136 aminotransferase K185A from acanthamoeba polyphaga mimivirus in the presence of the UDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-dihydroxy-5-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyloxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFQ
 
 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
9CJ0
 
 | |
9BGP
 
 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, aminotransferase | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
9BGR
 
 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its external aldimine with 2,3-diacetamido-4-amino-2,3,4-trideoxy-l-arabinose | | Descriptor: | (2R,3R,4R,5R)-3,4-diacetamido-5-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methoxy)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
7N63
 
 | | X-ray structure of HCAN_0200, an aminotransferase from Helicobacter canadensis in complex with its external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Griffiths, W.A, Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7B
 
 | | crystal structure of the N-formyltrasferase HCAN_0200 from Helicobacter canadensis on complex with folinic acid and dTDP-3-aminofucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7A
 
 | | crystal structure of the dTDP-Qui3N N-formyltransferase from Helicobacter canadensis, apo form | | Descriptor: | 1,2-ETHANEDIOL, Formyl_trans_N domain-containing protein | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7C
 
 | | crystal structure of the N-formyltransferase HCAN_0200 from helicobacter canadensis in complex with folinic acid and dTDP-3-aminoquinovose | | Descriptor: | 1,2-ETHANEDIOL, Formyl_trans_N domain-containing protein, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N67
 
 | | Crystal structure of HCAN_0198, a 3,4-ketoisomerase from Helicobacter canadensis | | Descriptor: | FdtA domain-containing protein, TETRAMETHYLAMMONIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Heisdorf, C.J, Griffiths, W.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
3TEA
 
 | | Crystal structure of Arthrobacter sp. strain su 4-hydroxybenzoyl CoA thioesterase double mutant Q58E/E73A complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
