1LRJ
 
 | | Crystal Structure of E. coli UDP-Galactose 4-Epimerase Complexed with UDP-N-Acetylglucosamine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
1M6V
 
 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
1MN0
 
 | | Crystal structure of galactose mutarotase from lactococcus lactis complexed with D-xylose | | Descriptor: | Aldose 1-epimerase, NICKEL (II) ION, alpha-D-xylopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1MMY
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-quinovose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-D-quinovopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1MMU
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-glucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-D-glucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1JXZ
 
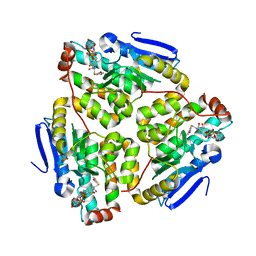 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
2GMU
 
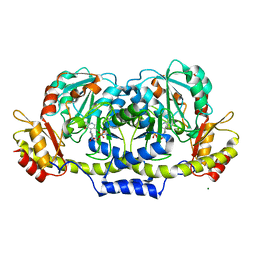 | | Crystal structure of E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase complexed with PLP-glutamate ketimine intermediate | | Descriptor: | MAGNESIUM ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, Putative pyridoxamine 5-phosphate-dependent dehydrase, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
5KTD
 
 | | FdhC with bound products: Coenzyme A and dTDP-3-amino-3,6-dideoxy-d-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
4ZTC
 
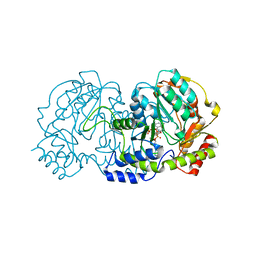 | | PglE Aminotransferase in complex with External Aldimine, Mutant K184A | | Descriptor: | Aminotransferase homolog, [(2R,3R,4R,5S,6R)-3-acetamido-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-oxidanyl-oxan-2-yl] [[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Riegert, A.S, Thoden, J.B, Young, N.M, Watson, D.C, Holden, H.M. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the external aldimine form of PglE, an aminotransferase required for N,N'-diacetylbacillosamine biosynthesis.
Protein Sci., 24, 2015
|
|
3M9V
 
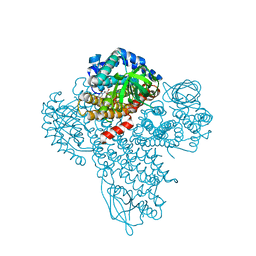 | |
7N63
 
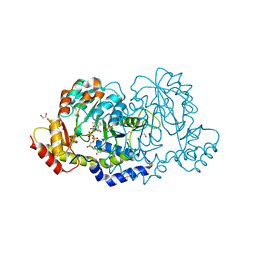 | | X-ray structure of HCAN_0200, an aminotransferase from Helicobacter canadensis in complex with its external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Griffiths, W.A, Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7B
 
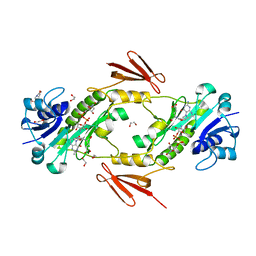 | | crystal structure of the N-formyltrasferase HCAN_0200 from Helicobacter canadensis on complex with folinic acid and dTDP-3-aminofucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7C
 
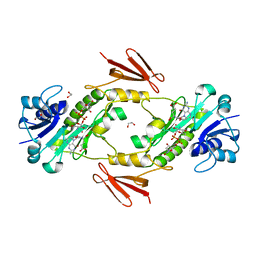 | | crystal structure of the N-formyltransferase HCAN_0200 from helicobacter canadensis in complex with folinic acid and dTDP-3-aminoquinovose | | Descriptor: | 1,2-ETHANEDIOL, Formyl_trans_N domain-containing protein, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7A
 
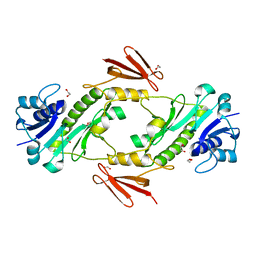 | | crystal structure of the dTDP-Qui3N N-formyltransferase from Helicobacter canadensis, apo form | | Descriptor: | 1,2-ETHANEDIOL, Formyl_trans_N domain-containing protein | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N67
 
 | | Crystal structure of HCAN_0198, a 3,4-ketoisomerase from Helicobacter canadensis | | Descriptor: | FdtA domain-containing protein, TETRAMETHYLAMMONIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Heisdorf, C.J, Griffiths, W.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
5VYT
 
 | | Crystal structure of the WbkC N-formyltransferase (F142A variant) from Brucella melitensis | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-mannose formyltransferase, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
5W71
 
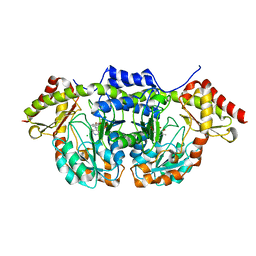 | | X-ray structure of BtrR from Bacillus circulans in the presence of the 2-DOS external aldimine | | Descriptor: | CHLORIDE ION, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5VYU
 
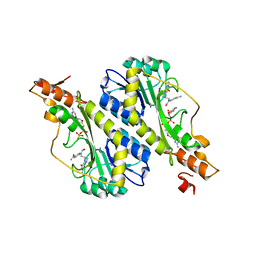 | | Crystal structure of the WbkC N-formyltransferase from Brucella melitensis in complex with GDP-perosaminea and N-10-formyltetrahydrofolate | | Descriptor: | GDP-perosamine, GUANOSINE-5'-DIPHOSPHATE, Gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-mannose formyltransferase, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
5VYQ
 
 | | Crystal structure of the N-formyltransferase Rv3404c from mycobacterium tuberculosis in complex with YDP-Qui4N and folinic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dunsirn, M.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Investigation of Rv3404c from Mycobacterium tuberculosis.
Biochemistry, 56, 2017
|
|
5W70
 
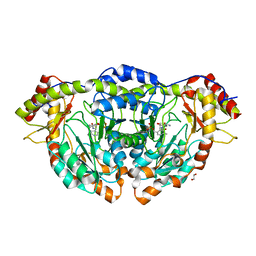 | | X-ray Structure of RbmB from Streptomyces ribosidificus | | Descriptor: | 1,2-ETHANEDIOL, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, [4-({[(1R,2S,3S,4R,5S)-5-amino-2,3,4-trihydroxycyclohexyl]amino}methyl)-5-hydroxy-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5VYR
 
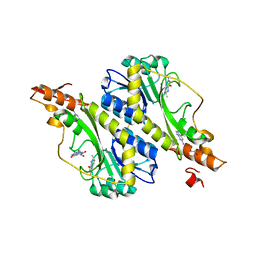 | | Crystal structure of the WbkC formyl transferase from Brucella melitensis | | Descriptor: | (6R)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
7S3W
 
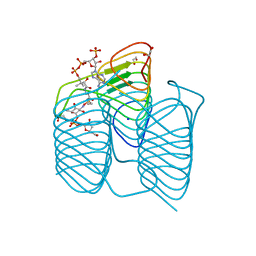 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S45
 
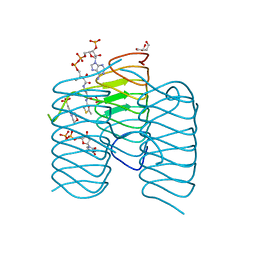 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Acetyl Coenzyme A and dTDP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S3U
 
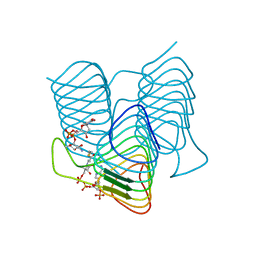 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S44
 
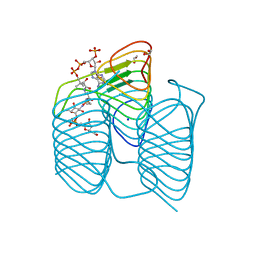 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
