7N4F
 
 | |
7N4G
 
 | |
8SJH
 
 | |
8SJG
 
 | |
8SJF
 
 | |
8SJI
 
 | |
5XZI
 
 | |
5XZJ
 
 | |
5WPN
 
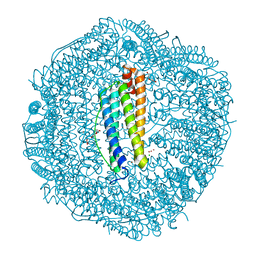 | | Zn-bound Structure of Chaetopterus variopedatus Ferritin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | De Meulenaere, E, Bailey, J.B, Tezcan, F.A, Deheyn, D. | | Deposit date: | 2017-08-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | First biochemical and crystallographic characterization of a fast-performing ferritin from a marine invertebrate.
Biochem. J., 474, 2017
|
|
5UP8
 
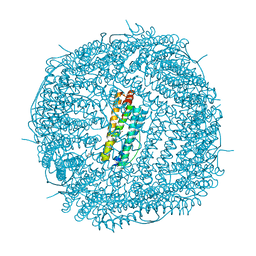 | | Crystal Structure of the Zn-bound Human Heavy-Chain variant 122H-delta C-star with para-benzenedihydroxamate | | Descriptor: | Ferritin heavy chain, N,N'-dihydroxybenzene-1,4-dicarboxamide, SODIUM ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.C, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5UP9
 
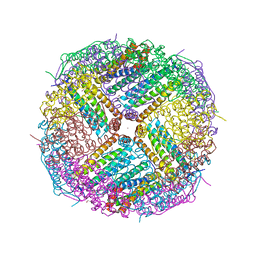 | | Crystal Structure of Zn-bound Human Heavy-Chain ferritin variant 122H-delta C-star with para-xylenedihydroxamate | | Descriptor: | 2,2'-(1,4-phenylene)bis(N-hydroxyacetamide), DI(HYDROXYETHYL)ETHER, Ferritin heavy chain, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5UP7
 
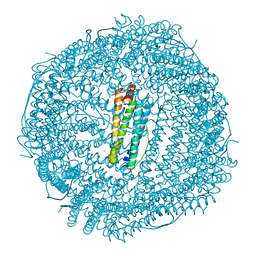 | | Crystal Structure of the Ni-bound Human Heavy-Chain Ferritin 122H-delta C-star variant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
7UT9
 
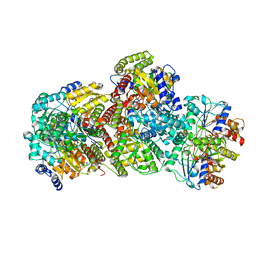 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ADP/ATP-bound) during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT8
 
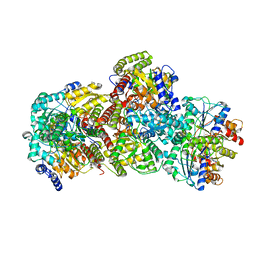 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ATP-bound) during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT6
 
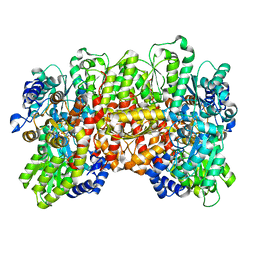 | | C1 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UTA
 
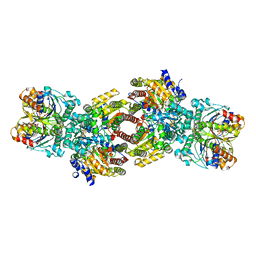 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (2:1 FeP:MoFeP) inhibited by BeFx during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM, ... | | Authors: | Rutledge, H.L, Cook, B.D, Nguyen, H.P.M, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT7
 
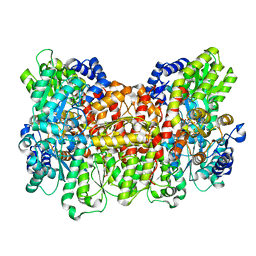 | | C2 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B.D, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
5KOJ
 
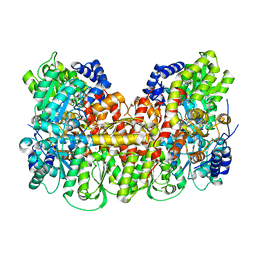 | | Nitrogenase MoFeP protein in the IDS oxidized state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Tezcan, F.A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
J.Am.Chem.Soc., 138, 2016
|
|
5KOH
 
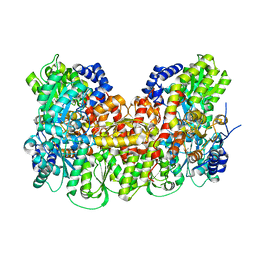 | | Nitrogenase MoFeP from Gluconacetobacter diazotrophicus in dithionite reduced state | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Owens, C.P, Tezcan, F.A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
J.Am.Chem.Soc., 138, 2016
|
|
5L32
 
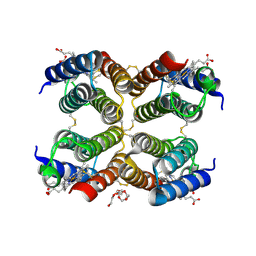 | |
5BU7
 
 | |
5CMQ
 
 | | Crystal Structure of Zn-bound Human H-Ferritin variant 122H-delta C-star | | Descriptor: | Ferritin heavy chain, ZINC ION | | Authors: | Sontz, P.A, Bailey, J.B, Ahn, S, Tezcan, F.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | A Metal Organic Framework with Spherical Protein Nodes: Rational Chemical Design of 3D Protein Crystals.
J.Am.Chem.Soc., 137, 2015
|
|
5CMR
 
 | | Crystal Structure of Linker-Mediated Zn-bound Human H-Ferritin variant 122H-delta C-star | | Descriptor: | Ferritin heavy chain, N,N'-dihydroxybenzene-1,4-dicarboxamide, SODIUM ION, ... | | Authors: | Sontz, P.A, Bailey, J.B, Ahn, S, Tezcan, F.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.792 Å) | | Cite: | A Metal Organic Framework with Spherical Protein Nodes: Rational Chemical Design of 3D Protein Crystals.
J.Am.Chem.Soc., 137, 2015
|
|
5CX1
 
 | | Nitrogenase molybdenum-iron protein beta-K400E mutant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Luca, M.A, Tezcan, F.A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7476 Å) | | Cite: | Evidence for Functionally Relevant Encounter Complexes in Nitrogenase Catalysis.
J.Am.Chem.Soc., 137, 2015
|
|
6O7S
 
 | |
