4EMY
 
 | |
6MGY
 
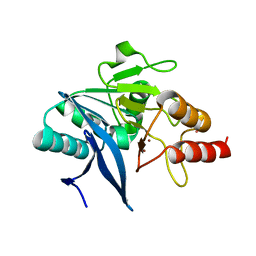 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae | | Descriptor: | GLYCEROL, Metallo-beta-lactamase NDM-5, POTASSIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae
To Be Published
|
|
3IC3
 
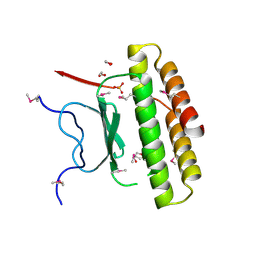 | | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Jedrzejczak, R, Mckinlay, J.B, Harwood, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009
TO BE PUBLISHED
|
|
3MLF
 
 | |
7JIW
 
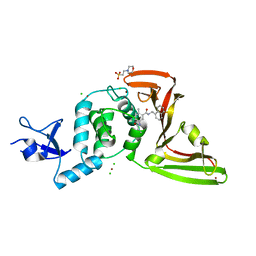 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIT
 
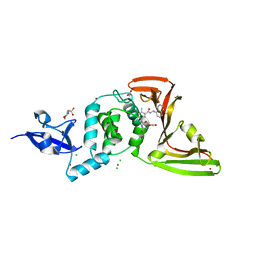 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
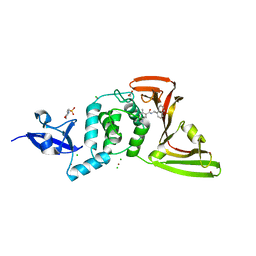 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7TRV
 
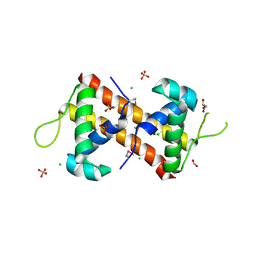 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TRW
 
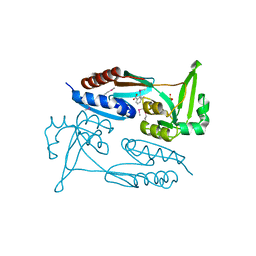 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | Authors: | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
3POC
 
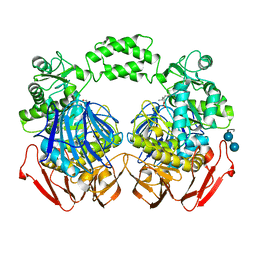 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
3GKX
 
 | |
3H1Q
 
 | | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ethanolamine utilization protein EutJ | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Kinney, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans
To be Published
|
|
3EA0
 
 | | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, ParA family, ... | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-24 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS
To be Published
|
|
3CP0
 
 | | Crystal structure of the soluble domain of membrane protein implicated in regulation of membrane protease activity from Corynebacterium glutamicum | | Descriptor: | CHLORIDE ION, Membrane protein implicated in regulation of membrane protease activity, ZINC ION | | Authors: | Kim, Y, Tesar, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-30 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the soluble domain of membrane protein implicated in regulation of membrane protease activity from Corynebacterium glutamicum.
To be Published
|
|
6OK0
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Sel1 repeat protein, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
3FN2
 
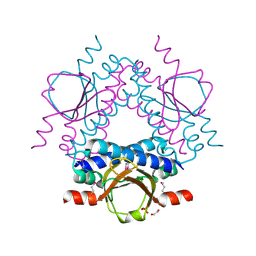 | | Crystal structure of a putative sensor histidine kinase domain from Clostridium symbiosum ATCC 14940 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a putative sensor histidine kinase domain from Clostridium symbiosum ATCC 14940
TO BE PUBLISHED
|
|
3FNC
 
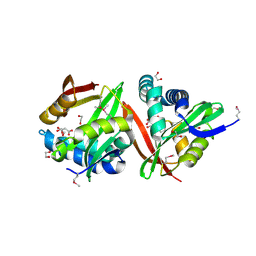 | | Crystal structure of a putative acetyltransferase from Listeria innocua | | Descriptor: | 1,2-ETHANEDIOL, MALONATE ION, Putative acetyltransferase | | Authors: | Cuff, M.E, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of a putative acetyltransferase from Listeria innocua.
TO BE PUBLISHED
|
|
3EWL
 
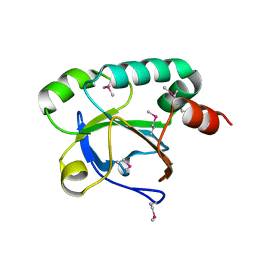 | |
3HTR
 
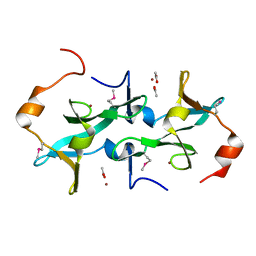 | | Crystal Structure of PRC-barrel Domain Protein from Rhodopseudomonas palustris | | Descriptor: | ACETIC ACID, ZINC ION, uncharacterized PRC-barrel Domain Protein | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Kinney, J, Babnigg, G, Harwood, C, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of PRC-barrel Domain Protein from Rhodopseudomonas palustris
To be Published
|
|
3FQ6
 
 | |
3KTB
 
 | | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482 | | Descriptor: | ACETIC ACID, Arsenical resistance operon trans-acting repressor, CALCIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-24 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482
To be Published
|
|
7SGV
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
7SGU
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor | | Descriptor: | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor
To Be Published
|
|
7SGW
 
 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
