1X4A
 
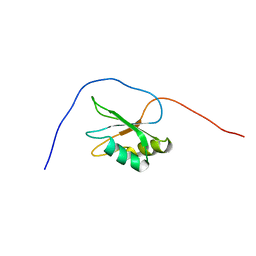 | | Solution structure of RRM domain in splicing factor SF2 | | Descriptor: | splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) variant | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor SF2
To be Published
|
|
1X4Q
 
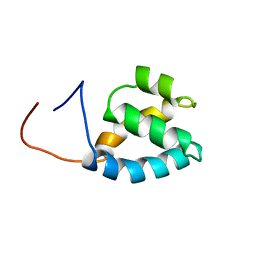 | | Solution structure of PWI domain in U4/U6 small nuclear ribonucleoprotein Prp3(hPrp3) | | Descriptor: | U4/U6 small nuclear ribonucleoprotein Prp3 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PWI domain in U4/U6 small nuclear ribonucleoprotein Prp3(hPrp3)
To be Published
|
|
1X47
 
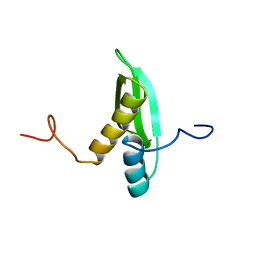 | | Solution structure of DSRM domain in DGCR8 protein | | Descriptor: | DGCR8 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DSRM domain in DGCR8 protein
To be Published
|
|
1X4P
 
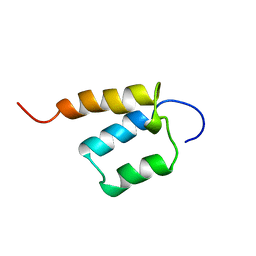 | | Solution structure of SURP domain in SFRS14 protei | | Descriptor: | Putative splicing factor, arginine/serine-rich 14 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in SFRS14 protei
To be Published
|
|
1X4I
 
 | | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3) | | Descriptor: | Inhibitor of growth protein 3, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3)
To be Published
|
|
2RQ4
 
 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-19 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
2ROK
 
 | | Solution structure of the cap-binding domain of PARN complexed with the cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, poly(A)-specific ribonuclease | | Authors: | Nagata, T, Suzuki, S, Endo, R, Shirouzu, M, Terada, T, Inoue, M, Kigawa, T, Guntert, P, Hayashizaki, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition.
Nucleic Acids Res., 36, 2008
|
|
2RQC
 
 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
1Z54
 
 | | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus | | Descriptor: | GLYCEROL, probable thioesterase | | Authors: | Ihsanawati, Kaminishi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus
TO BE PUBLISHED
|
|
1WDJ
 
 | | Crystal structure of TT1808 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1808 | | Authors: | Idaka, M, Wada, T, Murayama, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Of TT1808 From Thermus Thermophilus HB8
To be published
|
|
1WEP
 
 | | Solution structure of PHD domain in PHF8 | | Descriptor: | PHF8, ZINC ION | | Authors: | He, F, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in protein AA017385
To be Published
|
|
1X49
 
 | | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1X4N
 
 | | Solution structure of KH domain in FUSE binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in FUSE binding protein 1
To be Published
|
|
1X4R
 
 | | Solution structure of WWE domain in Parp14 protein | | Descriptor: | Parp14 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in Parp14 protein
To be Published
|
|
1X5S
 
 | | Solution structure of RRM domain in A18 hnRNP | | Descriptor: | Cold-inducible RNA-binding protein | | Authors: | Sato, A, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in A18 hnRNP
To be Published
|
|
1ZRJ
 
 | | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c | | Descriptor: | E1B-55kDa-associated protein 5 isoform c | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c
To be Published
|
|
2Z17
 
 | | Crystal structure of PDZ domain from human Pleckstrin homology, Sec7 | | Descriptor: | Pleckstrin homology Sec7 and coiled-coil domains-binding protein | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of PDZ domain from human Pleckstrin homology, Sec7
To be Published
|
|
1WEK
 
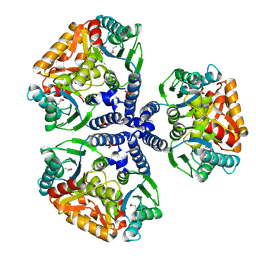 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
2YXM
 
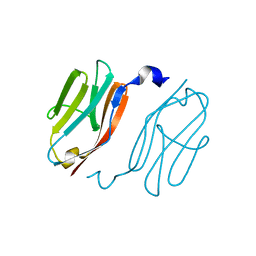 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YV8
 
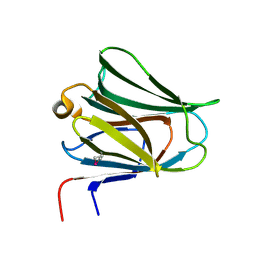 | | Crystal structure of N-terminal domain of human galectin-8 | | Descriptor: | Galectin-8 variant | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of N-terminal domain of human galectin-8
To be Published
|
|
2YY2
 
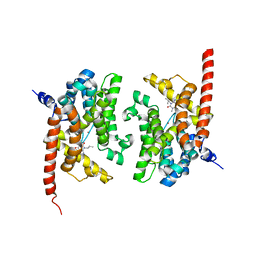 | | Crystal structure of the human Phosphodiesterase 9A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Handa, N, Shirouzu, M, Terada, T, Omori, K, Kotera, J, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human Phosphodiesterase 9A catalytic domain.
To be Published
|
|
2YXS
 
 | | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose | | Descriptor: | Galectin-8 variant, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose
To be Published
|
|
2Z0U
 
 | | Crystal structure of C2 domain of KIBRA protein | | Descriptor: | WW domain-containing protein 1 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C2 domain of KIBRA protein
To be Published
|
|
2Z0T
 
 | | Crystal structure of hypothetical protein PH0355 | | Descriptor: | Putative uncharacterized protein PH0355 | | Authors: | Murayama, K, Takemoto, C, Kato-Murayama, M, Kawazoe, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein PH0355
To be Published
|
|
2Z0I
 
 | | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus | | Descriptor: | Delta-aminolevulinic acid dehydratase | | Authors: | Wang, H, Xie, Y, Kawazoe, M, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus
To be Published
|
|
