1MQX
 
 | | NMR Solution Structure of Type-B Lantibiotics Mersacidin in MeOH/H2O Mixture | | 分子名称: | LANTIBIOTIC MERSACIDIN | | 著者 | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | 登録日 | 2002-09-17 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
1N1Q
 
 | | Crystal structure of a Dps protein from Bacillus brevis | | 分子名称: | DPS Protein, MU-OXO-DIIRON | | 著者 | Ren, B, Tibbelin, G, Kajino, T, Asami, O, Ladenstein, R. | | 登録日 | 2002-10-19 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Multi-layered Structure of Dps with a Novel Di-nuclear Ferroxidase Center
J.Mol.Biol., 329, 2003
|
|
1MQZ
 
 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | 分子名称: | LANTIBIOTIC MERSACIDIN | | 著者 | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | 登録日 | 2002-09-17 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
4V7G
 
 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | 著者 | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | 登録日 | 2009-09-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1MQY
 
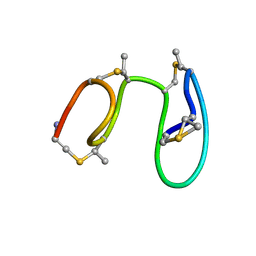 | | NMR solution structure of type-B lantibiotics mersacidin in DPC micelles | | 分子名称: | LANTIBIOTIC MERSACIDIN | | 著者 | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | 登録日 | 2002-09-17 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
2GEQ
 
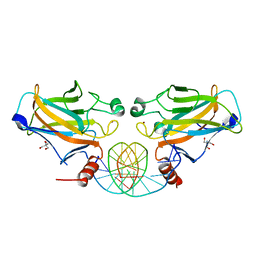 | | Crystal Structure of a p53 Core Dimer Bound to DNA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*GP*TP*GP*AP*GP*CP*AP*TP*GP*CP*TP*CP*AP*C)-3', Cellular tumor antigen p53, ... | | 著者 | Ho, W.C, Fitzgerald, M.X, Marmorstein, R. | | 登録日 | 2006-03-20 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the p53 Core Domain Dimer Bound to DNA.
J.Biol.Chem., 281, 2006
|
|
4PO2
 
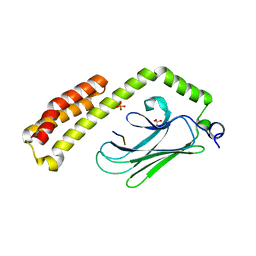 | | Crystal Structure of the Stress-Inducible Human Heat Shock Protein HSP70 Substrate-Binding Domain in Complex with Peptide Substrate | | 分子名称: | HSP70 substrate peptide, Heat shock 70 kDa protein 1A/1B, PHOSPHATE ION, ... | | 著者 | Zhang, P, Leu, J.I, Murphy, M.E, George, D.L, Marmorstein, R. | | 登録日 | 2014-02-24 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with Peptide substrate.
Plos One, 9, 2014
|
|
4PZT
 
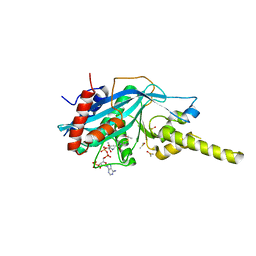 | | Crystal structure of p300 histone acetyltransferase domain in complex with an inhibitor, Acetonyl-Coenzyme A | | 分子名称: | DIMETHYL SULFOXIDE, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | 著者 | Maksimoska, J, Marmorstein, R. | | 登録日 | 2014-03-31 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the p300 Histone Acetyltransferase Bound to Acetyl-Coenzyme A and Its Analogues.
Biochemistry, 53, 2014
|
|
6OVH
 
 | | Cryo-EM structure of Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | 分子名称: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | 著者 | Golub, E, Subramanian, R.H, Yan, X, Alberstein, R.G, Tezcan, F.A. | | 登録日 | 2019-05-07 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-19 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
7OJM
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXIC CONDITIONS | | 分子名称: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | 著者 | Bui, S, Steiner, R.A. | | 登録日 | 2021-05-16 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
7OKZ
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL- QUINOLIN-4(1H)-ONE UNDER HYPEROXIC CONDITIONS | | 分子名称: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | 著者 | Bui, S, Steiner, R.A. | | 登録日 | 2021-05-18 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8A97
 
 | | ROOM TEMPERATURE CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) UNDER XENON PRESSURE (30 bar) | | 分子名称: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, D(-)-TARTARIC ACID, XENON | | 著者 | Bui, S, Prange, T, Steiner, R.A. | | 登録日 | 2022-06-27 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.897 Å) | | 主引用文献 | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
6POE
 
 | |
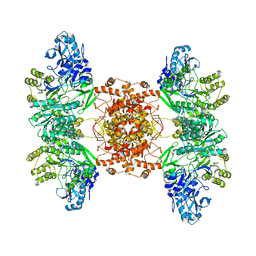
6POF
 
 | |
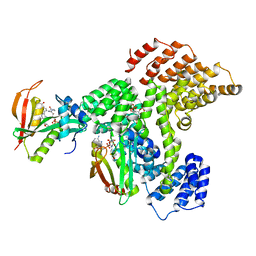
6PPL
 
 | |
6PW9
 
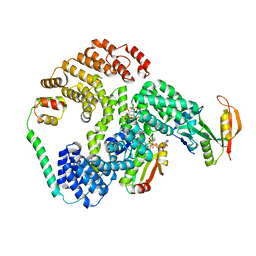 | | Cryo-EM structure of human NatE/HYPK complex | | 分子名称: | ACETYL COENZYME *A, Huntingtin-interacting protein K, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Deng, S, Marmorstein, R. | | 登録日 | 2019-07-22 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.03 Å) | | 主引用文献 | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
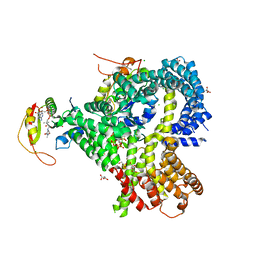
6O07
 
 | |
6EBC
 
 | | OhrB (Organic Hydroperoxide Resistance protein) wild type from Chromobacterium violaceum and reduced by DTT | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Organic hydroperoxide resistance protein | | 著者 | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | 登録日 | 2018-08-06 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EB3
 
 | | Structural and enzymatic characterization of an esterase from a metagenomic library | | 分子名称: | 1,2-ETHANEDIOL, 1,3-dihydroxypropan-2-yl butanoate, 2-hydroxypropane-1,3-diyl dibutanoate, ... | | 著者 | Guzzo, C.R, Carvalho, C.F, Teixeira, R.D, Farah, C.S. | | 登録日 | 2018-08-03 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Functional and Structural characterisation of an Esterase from Amazonian Dark Soil
To Be Published
|
|
6EBG
 
 | | Ohr (Organic Hydroperoxide Resistance protein) mutant - C60S interacting with dihydrolipoamide | | 分子名称: | (6S)-6,8-disulfanyloctanamide, Organic hydroperoxide resistance protein | | 著者 | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | 登録日 | 2018-08-06 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EBD
 
 | | OhrB (Organic Hydroperoxide Resistance protein) mutant (C60A) from Chromobacterium violaceum, interacting with dihydrolipoamide | | 分子名称: | (6S)-6,8-disulfanyloctanamide, CHLORIDE ION, Organic hydroperoxide resistance protein | | 著者 | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | 登録日 | 2018-08-06 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6ED0
 
 | |
6ECY
 
 | |
6EB4
 
 | | OhrB (Organic Hydroperoxide Resistance protein) from Chromobacterium violaceum | | 分子名称: | DI(HYDROXYETHYL)ETHER, Organic hydroperoxide resistance protein | | 著者 | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | 登録日 | 2018-08-04 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6FIZ
 
 | | Crystal Structure of CNG mimicking NaK-EAPP mutant (T67A) cocrystallized with K+ | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, POTASSIUM ION, ... | | 著者 | Napolitano, L.M.R, De March, M, Steiner, R.A, Onesti, S. | | 登録日 | 2018-01-19 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Crystal Structure of CNG mimicking NaK-EAPP mutant (T67A) cocrystallized with K+
To Be Published
|
|
