3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3VGI
 
 | | The crystal structure of hyperthermophilic family 12 endocellulase from Pyrococcus furiosus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Endoglucanase A, ... | | Authors: | Kataoka, M, Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-08-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus.
Febs Lett., 586, 2012
|
|
3WQ1
 
 | |
3WQ7
 
 | |
3WT3
 
 | |
3WY6
 
 | |
3WQ0
 
 | |
3WR0
 
 | |
3WXP
 
 | |
6AZ3
 
 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
4R07
 
 | | Crystal structure of human TLR8 in complex with ORN06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3'-O-[(R)-{[(2R,3aR,4R,6R,6aR)-6-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-hydroxy-2-oxidotetrahydrofuro[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]uridine 5'-(dihydrogen phosphate), ... | | Authors: | Tanji, H, Ohto, U, Shibata, T, Taoka, M, Yamauchi, Y, Isobe, T, Miyake, K, Shimizu, T. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toll-like receptor 8 senses degradation products of single-stranded RNA
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WH2
 
 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
3EQN
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | Descriptor: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
3EQO
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | Descriptor: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
6K0H
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-GlcNAc | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
6K0I
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-Glc | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
6K0G
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
4KTR
 
 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with isofagomine and glycerol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
4UNI
 
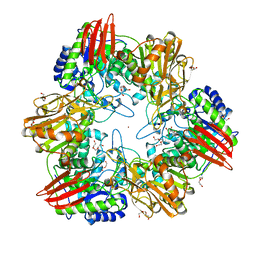 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOZ
 
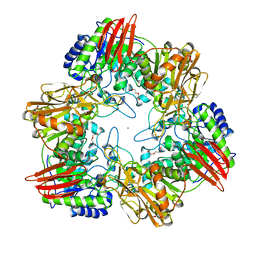 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4TMC
 
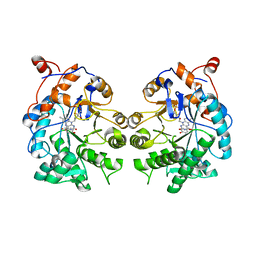 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
