3IEG
 
 | | Crystal Structure of P58(IPK) TPR Domain at 2.5 A | | Descriptor: | DnaJ homolog subfamily C member 3 | | Authors: | Tao, J, Sha, B. | | Deposit date: | 2009-07-22 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of P58(IPK) TPR Fragment Reveals the Mechanism for its Molecular Chaperone Activity in UPR.
J.Mol.Biol., 64, 2010
|
|
4GK0
 
 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
4GK5
 
 | | Crystal structure of human Rev3-Rev7-Rev1-Polkappa complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
6LNI
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5GS5
 
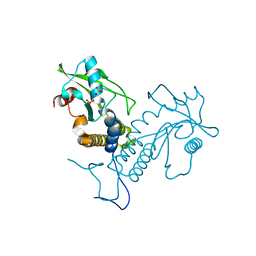 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
5GRM
 
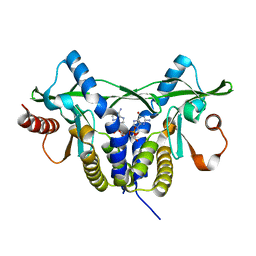 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
3IYM
 
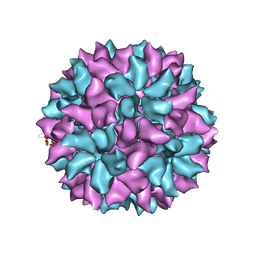 | | Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling | | Descriptor: | Capsid protein | | Authors: | Tang, J, Pan, J, Havens, W.F, Ochoa, W.F, Li, H, Sinkovits, R.S, Guu, T.S.Y, Ghabrial, S.A, Nibert, M.L, Tao, J.Y, Baker, T.S. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Backbone Trace of Partitivirus Capsid Protein from Electron Cryomicroscopy and Homology Modeling
Biophys.J., 99, 2010
|
|
3J1Z
 
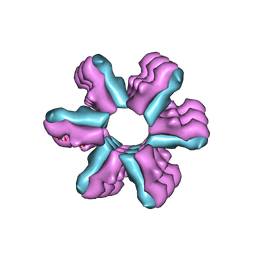 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7XYF
 
 | |
7XYG
 
 | |
1GAX
 
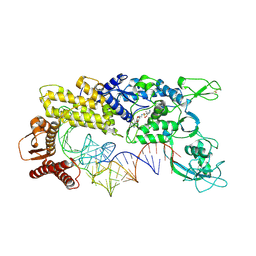 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, TRNA(VAL), VALYL-TRNA SYNTHETASE, ... | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Tao, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-06-23 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase.
Cell(Cambridge,Mass.), 103, 2000
|
|
7XXE
 
 | |
1APA
 
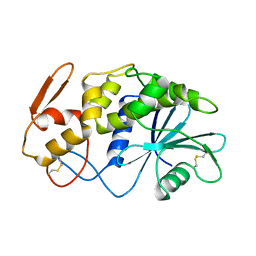 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
