1V3Z
 
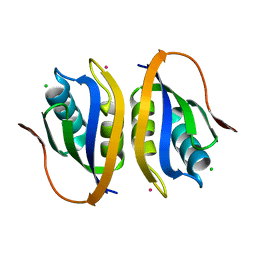 | |
1V5Y
 
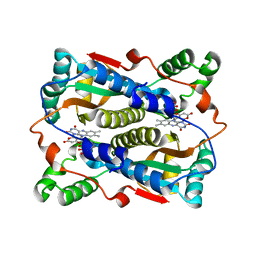 | | Binding of coumarins to NAD(P)H:FMN oxidoreductase | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, FLAVIN MONONUCLEOTIDE, Major NAD(P)H-flavin oxidoreductase | | Authors: | Kobori, T, Koike, H, Sasaki, H, Zenno, S, Saigo, K, Tanokura, M. | | Deposit date: | 2003-11-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of coumarins to NAD(P)H:FMN oxidoreductase
To be Published
|
|
2DE4
 
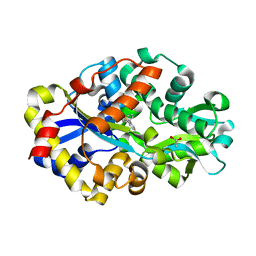 | | Crystal structure of DSZB C27S mutant in complex with biphenyl-2-sulfinic acid | | Descriptor: | 1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE2
 
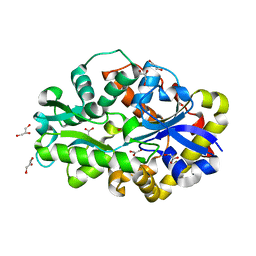 | | Crystal structure of desulfurization enzyme DSZB | | Descriptor: | ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, GLYCEROL | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE3
 
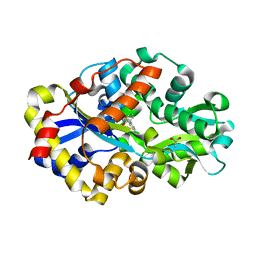 | | Crystal structure of DSZB C27S mutant in complex with 2'-hydroxybiphenyl-2-sulfinic acid | | Descriptor: | 2'-HYDROXY-1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, ... | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
3AAD
 
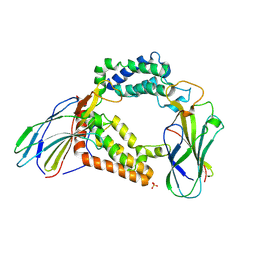 | | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, Transcription initiation factor TFIID subunit 1 | | Authors: | Akai, Y, Adachi, N, Hayashi, Y, Eitoku, M, Sano, N, Natsume, R, Kudo, N, Tanokura, M, Senda, T, Horikoshi, M. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZUA
 
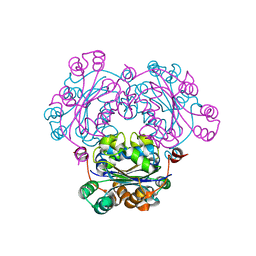 | | Crystal structure of nucleoside diphosphate kinase from Haloarcula quadrata | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Ichimura, T, Yamamura, A, Ohtsuka, J, Miyazono, K, Okai, M, Nagata, K, Tanokura, M. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular mechanism of distinct salt-dependent enzyme activity of two halophilic nucleoside diphosphate kinases
Biophys.J., 96, 2009
|
|
2Z1N
 
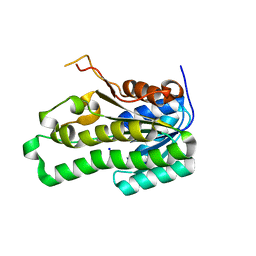 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | Descriptor: | SODIUM ION, dehydrogenase | | Authors: | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
3A76
 
 | | The crystal structure of LinA | | Descriptor: | GLYCEROL, Gamma-hexachlorocyclohexane dehydrochlorinase, SPERMIDINE | | Authors: | Okai, M, Kubota, K, Fukuda, M, Nagata, Y, Nagata, K, Tanokura, M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of g-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26
J.Mol.Biol., 2010
|
|
2YYS
 
 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
3WGB
 
 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WGC
 
 | | Aeromonas jandaei L-allo-threonine aldolase H128Y/S292R double mutant | | Descriptor: | L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2ZBC
 
 | | Crystal structure of STS042, a stand-alone RAM module protein, from hyperthermophilic archaeon Sulfolobus tokodaii strain7. | | Descriptor: | 83aa long hypothetical transcriptional regulator asnC, ISOLEUCINE | | Authors: | Miyazono, K, Tsujimura, M, Kawarabayasi, Y, Tanokura, M. | | Deposit date: | 2007-10-19 | | Release date: | 2008-03-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of STS042, a stand-alone RAM module protein, from hyperthermophilic archaeon Sulfolobus tokodaii strain7
Proteins, 71, 2008
|
|
3AI2
 
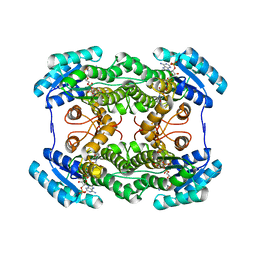 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI3
 
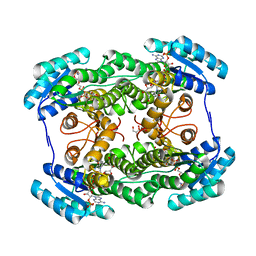 | | The crystal structure of L-Sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose | | Descriptor: | L-sorbose, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase, ... | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI1
 
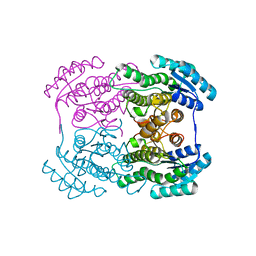 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | Descriptor: | NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3WLX
 
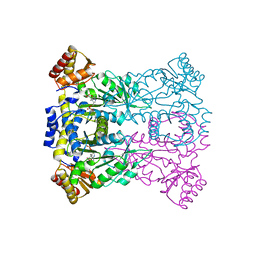 | | Crystal structure of low-specificity L-threonine aldolase from Escherichia coli | | Descriptor: | Low specificity L-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.-M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Hou, F, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure analysis of L-threonine aldolase from Escherichia coli unravels the low-specificity and thermostability
To be Published
|
|
3AK4
 
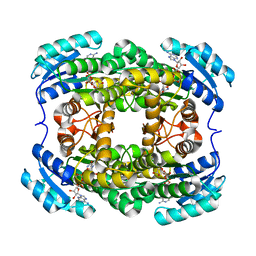 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
2YQY
 
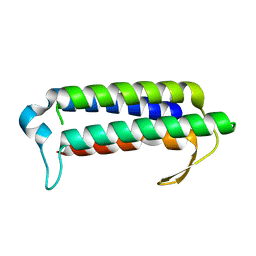 | | Crystal structure of TT2238, a four-helix bundle protein | | Descriptor: | Hypothetical protein TTHA0303 | | Authors: | Nagata, K, Ohtsuka, J, Iino, H, Ebihara, A, Yokoyama, S, Kuramitsu, S, Tanokura, M. | | Deposit date: | 2007-03-31 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TTHA0303 (TT2238), a four-helix bundle protein with an exposed histidine triad from Thermus thermophilus HB8 at 2.0 A
Proteins, 70, 2008
|
|
3W20
 
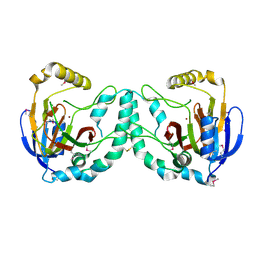 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3VXG
 
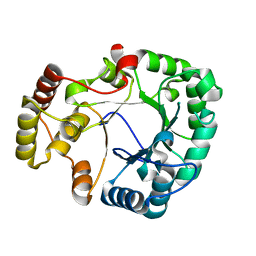 | | Crystal structure of conjugated polyketone reductase C2 from Candida Parapsilosis | | Descriptor: | Conjugated polyketone reductase C2 | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3VVU
 
 | | Crystal structure of reconstructed bacterial ancestral NDK, Bac1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VVT
 
 | | Crystal structure of reconstructed archaeal ancestral NDK, Arc1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W21
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3WI7
 
 | | Crystal Structure of the Novel Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
