6D97
 
 | |
6BSN
 
 | |
2RBF
 
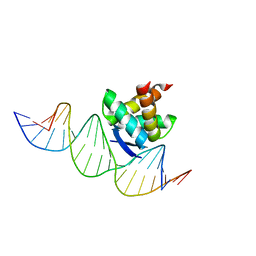 | | Structure of the ribbon-helix-helix domain of Escherichia coli PutA (PutA52) complexed with operator DNA (O2) | | Descriptor: | Bifunctional protein putA, DNA (5'-D(*DTP*DT*DTP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DCP*DAP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DGP*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DAP*DAP*DA)-3') | | Authors: | Tanner, J.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
J.Mol.Biol., 381, 2008
|
|
7MYA
 
 | |
7MYC
 
 | |
4ZVW
 
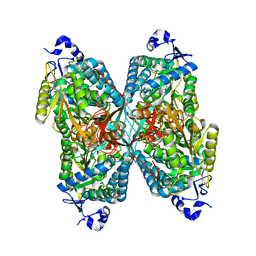 | | Structure of apo human ALDH7A1 in space group C2 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
4ZVY
 
 | |
4ZVX
 
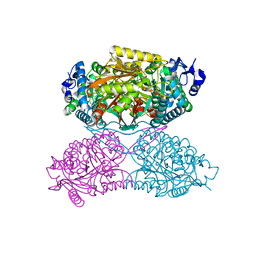 | | Structure of apo human ALDH7A1 in space group P4212 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
5CDH
 
 | |
5CKU
 
 | |
1TJ1
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1XVJ
 
 | | Crystal Structure Of Rat alpha-Parvalbumin D94S/G98E Mutant | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Tanner, J.J, Agah, S, Lee, Y.H, Henzl, M.T. | | Deposit date: | 2004-10-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the D94S/G98E Variant of Rat alpha-Parvalbumin. An Explanation for the Reduced Divalent Ion Affinity.
Biochemistry, 44, 2005
|
|
1XKJ
 
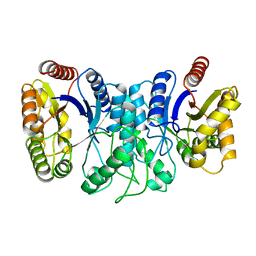 | | BACTERIAL LUCIFERASE BETA2 HOMODIMER | | Descriptor: | BETA2 LUCIFERASE | | Authors: | Tanner, J.J, Krause, K.L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial luciferase beta 2 homodimer: implications for flavin binding.
Biochemistry, 36, 1997
|
|
1TJ2
 
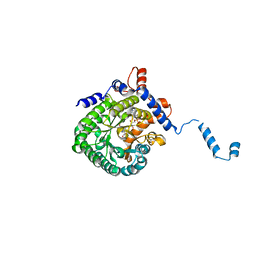 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | Descriptor: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
3E2Q
 
 | |
4O8A
 
 | |
3E2S
 
 | | Crystal Structure Reduced PutA86-630 Mutant Y540S Complexed with L-proline | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PENTAETHYLENE GLYCOL, PROLINE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2008-08-06 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate.
Biochemistry, 48, 2009
|
|
7NA0
 
 | |
7MYB
 
 | |
7MY9
 
 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Campbell, A.C. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MWT
 
 | |
7MWV
 
 | |
7MWU
 
 | |
3E2R
 
 | |
3ET4
 
 | | Structure of Recombinant Haemophilus Influenzae E(P4) Acid Phosphatase | | Descriptor: | MAGNESIUM ION, Outer membrane protein P4, NADP phosphatase, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Recombinant Haemophilus Influenzae E (P4) Acid Phosphatase
Reveals a New Member of the Haloacid Dehalogenase Superfamily.
Biochemistry, 46, 2007
|
|
