7FCW
 
 | | X-ray structure of H2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FCU
 
 | | X-ray structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3VZR
 
 | |
1WLT
 
 | | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue from Sulfolobus tokodaii | | Descriptor: | 176aa long hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Watanabe, N, Sakai, N, Zilian, Z, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue
to be published
|
|
1WMI
 
 | | Crystal structure of archaeal RelE-RelB complex from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PHS013, hypothetical protein PHS014 | | Authors: | Takagi, H, Kakuta, Y, Kamachi, R, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects
Nat.Struct.Mol.Biol., 12, 2005
|
|
1VE0
 
 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
1UC2
 
 | | Hypothetical Extein Protein of PH1602 from Pyrococcus horikoshii | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, hypothetical protein PH1602 | | Authors: | Okada, C, Maegawa, Y, Yao, M, Tanaka, I. | | Deposit date: | 2003-04-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold
Proteins, 63, 2006
|
|
3X2P
 
 | | Neutron and X-ray joint refined structure of PcCel45A with cellopentaose at 298K. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.518 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2O
 
 | | Neutron and X-ray joint refined structure of PcCel45A apo form at 298K. | | Descriptor: | Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
1UCD
 
 | | Crystal structure of Ribonuclease MC1 from bitter gourd seeds complexed with 5'-UMP | | Descriptor: | Ribonuclease MC, URACIL, URIDINE-5'-MONOPHOSPHATE | | Authors: | Suzuki, A, Numata, T, Yao, M, Kimura, M, Tanaka, I. | | Deposit date: | 2003-04-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of RNase MC1 from bitter gourd seeds in complex with 5'UMP
To be published
|
|
1UDD
 
 | | TenA homologue protein from P.horikoshii OT3 | | Descriptor: | transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure analysis of PH1161 protein, a transcriptional activator TenA homologue from the hyperthermophilic archaeon Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UB9
 
 | | Structure of the transcriptional regulator homologue protein from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH1061 | | Authors: | Okada, U, Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the transcriptional regulator homolog protein from Pyrococcus horikoshii OT3.
Proteins, 63, 2006
|
|
2E52
 
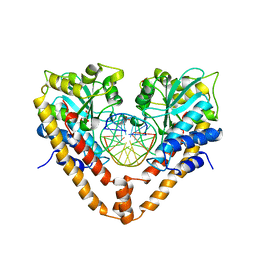 | | Crystal structural analysis of HindIII restriction endonuclease in complex with cognate DNA at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, DNA (5'-D(*DGP*DCP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DGP*DGP*DC)-3'), GLYCEROL, ... | | Authors: | Watanabe, N, Sato, C, Tanaka, I. | | Deposit date: | 2006-12-18 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1UIZ
 
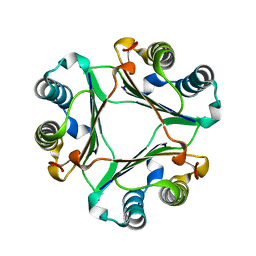 | | Crystal Structure Of Macrophage Migration Inhibitory Factor From Xenopus Laevis. | | Descriptor: | Macrophage Migration Inhibitory Factor | | Authors: | Suzuki, M, Takamura, Y, Maeno, M, Tochinai, S, Iyaguchi, D, Tanaka, I, Nishihira, J, Ishibashi, T. | | Deposit date: | 2003-07-24 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenopus laevis Macrophage Migration Inhibitory Factor Is Essential for Axis Formation and Neural Development.
J.Biol.Chem., 279, 2004
|
|
1VAJ
 
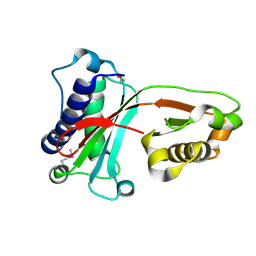 | | Crystal Structure of Uncharacterized Protein PH0010 From Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0010 | | Authors: | Tajika, Y, Sakai, N, Tamura, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-02-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of PH0010 from Pyrococcus horikoshii, which is highly homologous to human AMMECR 1C-terminal region
Proteins, 58, 2005
|
|
1V9G
 
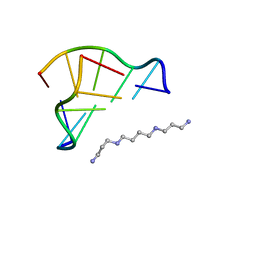 | | Neutron Crystallographic analysis of the Z-DNA hexamer CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', N,N'-BIS(3-AMMONIOPROPYL)BUTANE-1,4-DIAMINIUM | | Authors: | Chatake, T, Tanaka, I, Niimura, N. | | Deposit date: | 2004-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ULY
 
 | | Crystal structure analysis of the ArsR homologue DNA-binding protein from P. horikoshii OT3 | | Descriptor: | hypothetical protein PH1932 | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-09-17 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the PH1932 protein, a unique archaeal ArsR type winged-HTH transcription factor from Pyrococcus horikoshii OT3
Proteins, 70, 2008
|
|
1V7O
 
 | |
3A8E
 
 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1V43
 
 | | Crystal Structure of ATPase subunit of ABC Sugar Transporter | | Descriptor: | sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-11-08 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
1V30
 
 | | Crystal Structure Of Uncharacterized Protein PH0828 From Pyrococcus horikoshii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical UPF0131 protein PH0828 | | Authors: | Tajika, Y, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-10-21 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of hypothetical protein PH0828 from Pyrococcus horikoshii.
Proteins, 57, 2004
|
|
1V7L
 
 | |
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UCC
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
