2ZCB
 
 | | Crystal Structure of ubiquitin P37A/P38A | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Kitahara, R, Tanaka, T, Sakata, E, Yamaguchi, Y, Kato, K, Yokoyama, S. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of ubiquitin P37A/P38A
To be published
|
|
5B22
 
 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
1GLV
 
 | |
1IPB
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
2IV4
 
 | | hPrP180-195 structure | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Saviano, G, Tancredi, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Conformational Landscape of the Human Prion Protein Alpha 2 Domain: Comparative NMR and Md Studies on Helix-2-Derived Peptides
To be Published
|
|
2IV5
 
 | | hPrP-173-195 solution structure | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Saviano, G, Tancredi, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Conformational Landscape of the Human Prion Protein Alpha 2 Domain: Comparative NMR and Md Studies on Helix-2-Derived Peptides
To be Published
|
|
3W1D
 
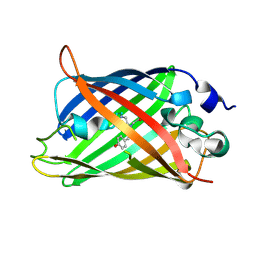 | |
3W1C
 
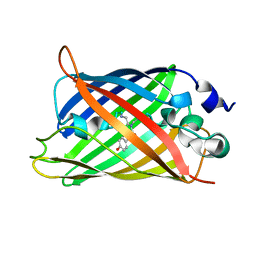 | |
3WXC
 
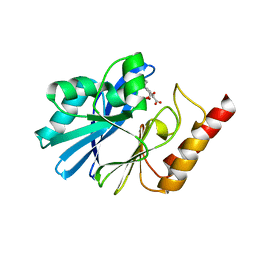 | | Crystal Structure of IMP-1 metallo-beta-lactamase complexed with a 3-aminophtalic acid inhibitor | | Descriptor: | 3-(4-hydroxypiperidin-1-yl)benzene-1,2-dicarboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Saito, J, Watanabe, T, Yamada, M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic analysis of IMP-1 metallo-beta-lactamase complexed with a 3-aminophthalic acid derivative, structure-based drug design, and synthesis of 3,6-disubstituted phthalic acid derivative inhibitors
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2IV6
 
 | | hPrP-173-195-D178N solution structure | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Saviano, G, Tancredi, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Conformational Landscape of the Human Prion Protein Alpha 2 Domain: Comparative NMR and Md Studies on Helix-2-Derived Peptides
To be Published
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
1QWQ
 
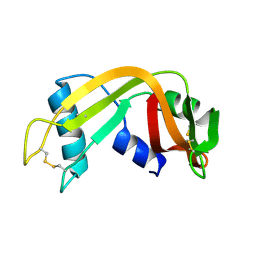 | | Solution structure of the monomeric N67D mutant of Bovine Seminal Ribonuclease | | Descriptor: | Ribonuclease | | Authors: | Avitabile, F, Alfano, C, Spadaccini, R, Crescenzi, O, D'Ursi, A.M, D'Alessio, G, Tancredi, T, Picone, D. | | Deposit date: | 2003-09-03 | | Release date: | 2003-09-16 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | THE SWAPPING OF TERMINAL ARMS IN RIBONUCLEASES: COMPARISON OF THE SOLUTION STRUCTURE OF MONOMERIC BOVINE SEMINAL AND PANCREATIC RIBONUCLEASES
Biochemistry, 42, 2003
|
|
7CC4
 
 | |
1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
2GVS
 
 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
1VBL
 
 | | Structure of the thermostable pectate lyase PL 47 | | Descriptor: | CALCIUM ION, pectate lyase 47 | | Authors: | Nakaniwa, T, Tada, T, Yamaguchi, A, Kitatani, T, Takao, M, Sakai, T, Nishimura, K. | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Thermostability of Pectate Lyase from Bacillus sp. TS 47
To be Published
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1WVU
 
 | |
1JOY
 
 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
1U24
 
 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U25
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate in the C2221 crystal form | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1JSA
 
 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | Descriptor: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | Authors: | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
1U26
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
3WSU
 
 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
