7DNM
 
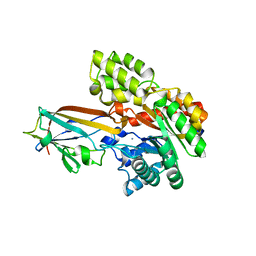 | | Crystal structure of the AgCarB2-C2 complex | | 分子名称: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | 著者 | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | 登録日 | 2020-12-10 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7TJ0
 
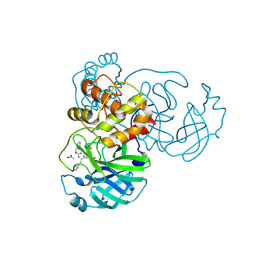 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7CK5
 
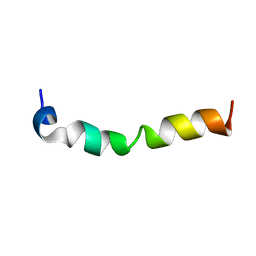 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | 分子名称: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | 著者 | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | 登録日 | 2020-07-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
7DVE
 
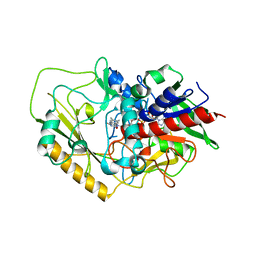 | | Crystal structure of FAD-dependent C-glycoside oxidase | | 分子名称: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | 登録日 | 2021-01-13 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BUW
 
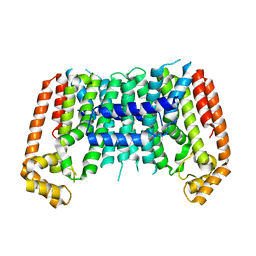 | | Eucommia ulmoides TPT3 mutant -C94Y/A95F | | 分子名称: | FPS3 | | 著者 | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | 登録日 | 2020-04-08 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUV
 
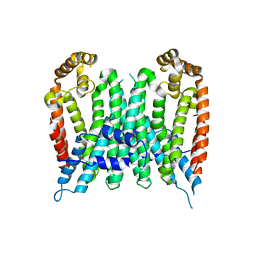 | | Eucommia ulmoides TPT3, crystal form 2 | | 分子名称: | FPS3 | | 著者 | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | 登録日 | 2020-04-08 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUU
 
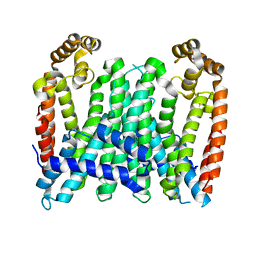 | | Eucommia ulmoides TPT3, crystal form 1 | | 分子名称: | FPS3 | | 著者 | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | 登録日 | 2020-04-08 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
3VND
 
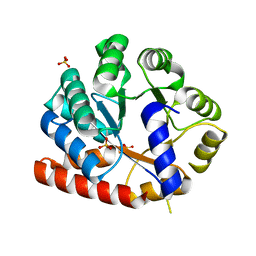 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | 著者 | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | 登録日 | 2012-01-12 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
2QKE
 
 | |
1U9I
 
 | | Crystal Structure of Circadian Clock Protein KaiC with Phosphorylation Sites | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, KaiC, MAGNESIUM ION | | 著者 | Xu, Y, Mori, T, Pattanayek, R, Pattanayek, S, Egli, M, Johnson, C.H. | | 登録日 | 2004-08-09 | | 公開日 | 2005-04-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
6JNJ
 
 | |
6JNK
 
 | |
7TJB
 
 | |
7TLV
 
 | |
7TRR
 
 | |
6L06
 
 | |
6L07
 
 | |
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | 著者 | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | 登録日 | 2022-09-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | 著者 | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | 登録日 | 2022-09-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | 著者 | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | 登録日 | 2022-09-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
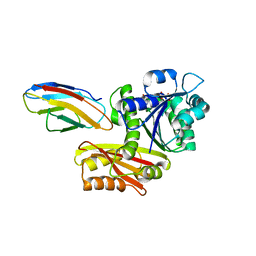 | | Escherichia coli FtsZ complexed with monobody (P21) | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | 著者 | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | 登録日 | 2022-09-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8H1O
 
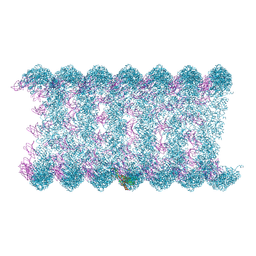 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | 著者 | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | 登録日 | 2022-10-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
7E7S
 
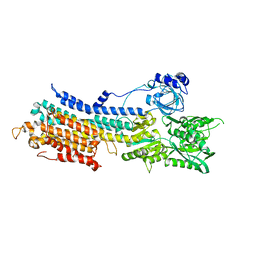 | | WT transporter state1 | | 分子名称: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | 著者 | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | 登録日 | 2021-02-27 | | 公開日 | 2021-09-08 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
8F3A
 
 | | HIV-1 gp41 coiled-coil pocket IQN17 | | 分子名称: | ACETIC ACID, CHLORIDE ION, IQN17 | | 著者 | Bruun, T.U.J, Tang, S, Fernandez, D, Kim, P.S. | | 登録日 | 2022-11-09 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-guided stabilization improves the ability of the HIV-1 gp41 hydrophobic pocket to elicit neutralizing antibodies.
J.Biol.Chem., 299, 2023
|
|
8F3B
 
 | |
