7C0D
 
 | |
7BYW
 
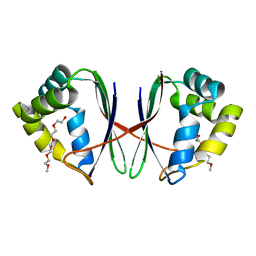 | |
7BYU
 
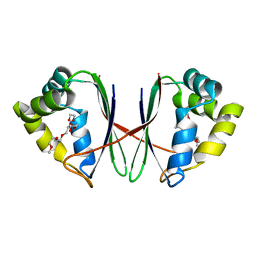 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7C0E
 
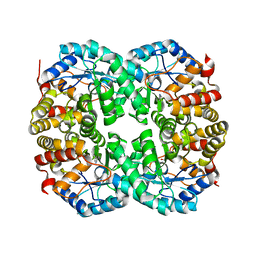 | |
7C0C
 
 | |
3WN6
 
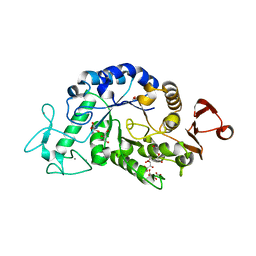 | | Crystal structure of alpha-amylase AmyI-1 from Oryza sativa | | Descriptor: | Alpha-amylase, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Ochiai, A, Sugai, H, Harada, K, Tanaka, S, Ishiyama, Y, Ito, K, Tanaka, T, Uchiumi, T, Taniguchi, M, Mitsui, T. | | Deposit date: | 2013-12-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of alpha-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability
Biosci.Biotechnol.Biochem., 78, 2014
|
|
7Q8T
 
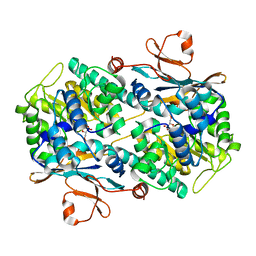 | | Crystal structure of NAMPT bound to ligand TSY535(compound 9a) | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION, [(2~{R},3~{S},4~{R},5~{S})-3,4-bis(oxidanyl)-5-[4-[[[4-(phenylsulfonyl)phenyl]carbamoylamino]methyl]phenyl]oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Kraemer, A, Tang, S, Butterworth, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Chemistry-led investigations into the mode of action of NAMPT activators, resulting in the discovery of non-pyridyl class NAMPT activators.
Acta Pharm Sin B, 13, 2023
|
|
3K09
 
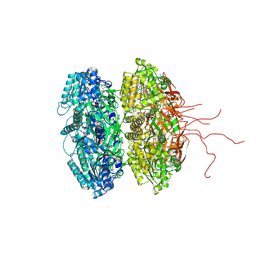 | | Crystal structure of the phosphorylation-site mutant S431D of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3JZM
 
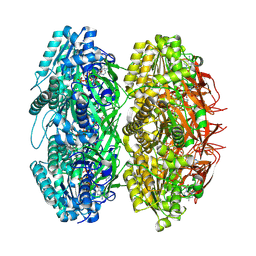 | | Crystal structure of the phosphorylation-site mutant T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-23 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0A
 
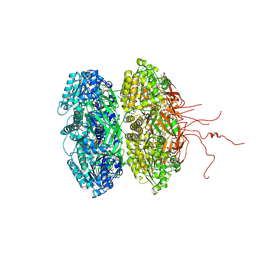 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0F
 
 | | Crystal structure of the phosphorylation-site double mutant T426A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0C
 
 | | Crystal structure of the phosphorylation-site double mutant S431A/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
6KSL
 
 | | Staphylococcus aureus lipase - S116A inactive mutant | | Descriptor: | CALCIUM ION, LAURIC ACID, Lipase 2, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
5YY0
 
 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
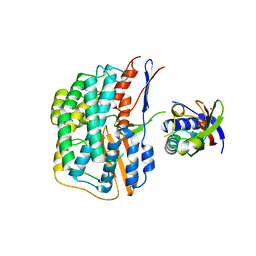 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2DJ4
 
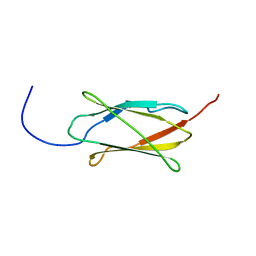 | | Solution structure of the 13th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 13th filamin domain from human Filamin-B
To be Published
|
|
2DHZ
 
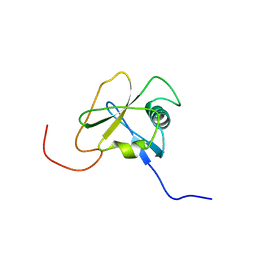 | | Solution Structure of the RA Domain in the Human Link Guanine Nucleotide Exchange Factor II (Link-GEFII) | | Descriptor: | Rap guanine nucleotide exchange factor (GEF)-like 1 | | Authors: | Zhao, C, Kigawa, T, Yoneyama, M, Koshiba, S, Harada, T, Watanabe, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RA Domain in the Human Link Guanine Nucleotide Exchange Factor II (Link-GEFII)
To be Published
|
|
2DS4
 
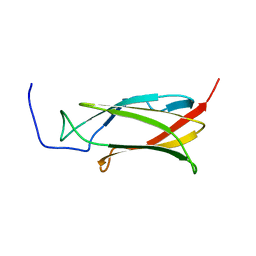 | | Solution structure of the filamin domain from human tripartite motif protein 45 | | Descriptor: | Tripartite motif protein 45 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-21 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the filamin domain from human tripartite motif protein 45
To be Published
|
|
2DIC
 
 | | Solution structure of the 12th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-29 | | Release date: | 2006-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 12th filamin domain from human Filamin-B
To be Published
|
|
2DMB
 
 | | Solution structure of the 15th Filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 15th Filamin domain from human Filamin-B
To be Published
|
|
2DI9
 
 | | Solution structure of the 9th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-29 | | Release date: | 2006-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 9th filamin domain from human Filamin-B
To be Published
|
|
2DI0
 
 | | Solution Structure of the CUE Domain in the Human Activating Signal Cointegrator 1 Complex Subunit 2 (ASCC2) | | Descriptor: | Activating signal cointegrator 1 complex subunit 2 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the CUE Domain in the Human Activating Signal Cointegrator 1 Complex Subunit 2 (ASCC2)
To be Published
|
|
2DHK
 
 | | Solution structure of the PH domain of TBC1 domain family member 2 protein from human | | Descriptor: | TBC1 domain family member 2 | | Authors: | Li, H, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2006-09-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of TBC1 domain family member 2 protein from human
To be Published
|
|
2DKL
 
 | | Solution Structure of the UBA Domain in the Human Trinucleotide Repeat Containing 6C Protein (hTNRC6C) | | Descriptor: | Trinucleotide Repeat Containing 6C Protein | | Authors: | Zhao, C, Kigawa, T, Yoneyama, M, Koshiba, S, Harada, T, Watanabe, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-12 | | Release date: | 2006-10-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the UBA Domain in the Human Trinucleotide Repeat Containing 6C Protein (hTNRC6C)
To be Published
|
|
2DLG
 
 | | Solution structure of the 20th Filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-18 | | Release date: | 2006-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 20th Filamin domain from human Filamin-B
To be Published
|
|
