2VP8
 
 | | Structure of Mycobacterium tuberculosis Rv1207 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPTEROATE SYNTHASE 2 | | Authors: | Gengenbacher, M, Xu, T, Niyomwattanakit, P, Spraggon, G, Dick, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biochemical and Structural Characterization of the Putative Dihydropteroate Synthase Ortholog Rv1207 of Mycobacterium Tuberculosis.
Fems Microbiol.Lett., 287, 2008
|
|
2RIV
 
 | | Crystal structure of the reactive loop cleaved human Thyroxine Binding Globulin | | Descriptor: | GLYCEROL, SULFATE ION, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
2K1E
 
 | | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA | | Descriptor: | water soluble analogue of potassium channel, KcsA | | Authors: | Ma, D, Xu, Y, Tillman, T, Tang, P, Meirovitch, E, Eckenhoff, R, Carnini, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3UBI
 
 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
4AXK
 
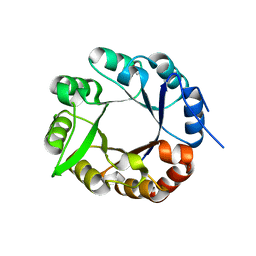 | | CRYSTAL STRUCTURE OF subHisA from the thermophile Corynebacterium efficiens | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, GLYCEROL | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Medina-Ruiz, S, Verduzco-Castro, E.A, Gaytan, P, Carrillo-Tripp, M, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2012-06-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of Substrate Specificity in a Recipient'S Enzyme Following Horizontal Gene Transfer.
Mol.Biol.Evol., 30, 2013
|
|
1VRY
 
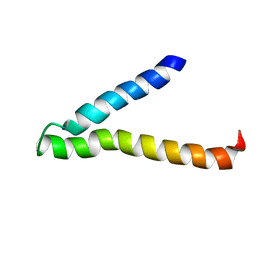 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
4LZF
 
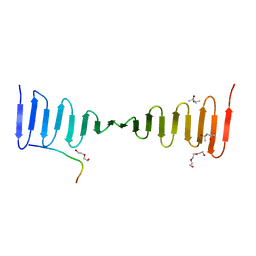 | | A novel domain in the microcephaly protein CPAP suggests a role in centriole architecture | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Centrosomal P4.1-associated protein, SCL-interrupting locus protein homolog, ... | | Authors: | Hatzopoulos, G.N, Erat, M.C, Cutts, E, Rogala, K, Slatter, L, Stansfeld, P.J, Vakonakis, I. | | Deposit date: | 2013-07-31 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture.
Structure, 21, 2013
|
|
2MHL
 
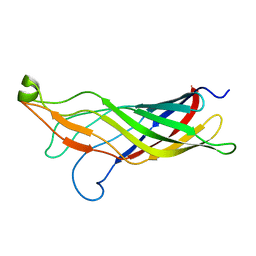 | |
2XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
2JGB
 
 | | Structure of human eIF4E homologous protein 4EHP with m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
2JGC
 
 | | Structure of the human eIF4E homologous protein, 4EHP without ligand bound | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
1C2Y
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PROTEIN (LUMAZINE SYNTHASE) | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-07-27 | | Release date: | 2000-07-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly.
Protein Sci., 8, 1999
|
|
3ZH9
 
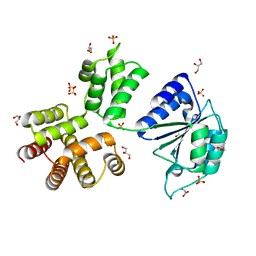 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
2K18
 
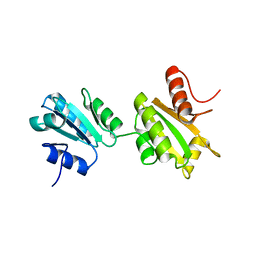 | | Solution structure of bb' domains of human protein disulfide isomerase | | Descriptor: | Protein disulfide-isomerase | | Authors: | Denisov, A.Y, Maattanen, P, Dabrowski, C, Kozlov, G, Thomas, D.Y, Gehring, K. | | Deposit date: | 2008-02-22 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the bb' domains of human protein disulfide isomerase.
Febs J., 276, 2009
|
|
1C41
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, LUMAZINE SYNTHASE, SULFATE ION | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly
Protein Sci., 8, 1999
|
|
3ZUS
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC-A-SNAP23-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN 23, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZUR
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC0-A-SNAP25-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZUQ
 
 | | Crystal structure of an engineered botulinum neurotoxin type B - derivative, LC-B-GS-Hn-B | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Y1W
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | 2-{4-[3-FLUORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL] PIPERIDIN-1-YL}-N-METHYLETHANAMINE, HISTONE-ARGININE METHYLTRANSFERASE CARM1, SINEFUNGIN | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
2Y1X
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | CHLORIDE ION, HISTONE-ARGININE METHYLTRANSFERASE CARM1, N-(3-{5-[5-(1H-INDOL-4-YL)-1,3,4-OXADIAZOL-2-YL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL}BENZYL)-L-ALANINAMIDE, ... | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
